Transposons families/Tn402 family: Difference between revisions
| (43 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
== Historical == | |||
Tn''402'' was first identified <ref> | [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] was first identified <ref name=":7">{{#pmid:321437}}</ref> as an insertion of [[wikipedia:Trimethoprim|trimethoprim]] resistance from the IncP plasmid R751 <ref>{{#pmid:4599661}}</ref><ref>Jacob A, Shapiro J, Yamamoto L, Smith DI, Cohen SN, Berg D. . Plasmids studied in Escherichia coli and other enteric bacteria. In (ed.),. In: Bukhari AI, Shapiro J, Adhya S, editors. DNA insertion elements, episomes and plasmids . Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.; 1977.</ref> into a [[wikipedia:Lambda_phage|bacteriophage lambda]] derivative phage. Historically, this family of Tn have been intimately linked to integrons and, indeed, [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']-like transposons have proved instrumental in integron dispersal. This first became apparent during the study of integron structures which include an integron integrase gene, ''intI'', and various antibiotic resistance gene cassettes. These structures were found in different sequence environments suggesting that they are mobile genetic elements. Since they did not contain typical transposition-related genes or obvious terminal inverted repeats, it was suggested that integrons were unusual mobile elements distinct from typical transposons <ref>{{#pmid:2560119}}</ref>. However, identification of an integron structure which is embedded in the [[Transposons families/Tn3 family|Tn''3'' family]] transposon, [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21''], and flanked by 25bp '''I'''nverted '''R'''epeats (IR), in turn flanked by 5bp '''D'''irect '''R'''epeats (DR), suggested that the integron was part of a precursor transposon <ref>{{#pmid:3007931}}</ref><ref name=":20">{{#pmid:1963947}}</ref> whose transposition genes may have decayed following insertion into [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21'']. | ||
Indeed, integrons In0, In2, and In5 were later identified as defective transposon derivatives <ref> | Indeed, integrons [https://tncentral.ncc.unesp.br/report/te/In0-U49101 In0], [https://tncentral.ncc.unesp.br/report/te/In2-AF071413 In2], and In5 were later identified as defective transposon derivatives <ref>{{#pmid:PMC178208}}</ref>. This clearly raises the question of how to define and name integrons (see [[Transposons families/Tn402 family#Origin of the Tn402 group integrons.|Integrons]]). We will use a working definition for an integron as a DNA segment composed of an “integron platform” together with a variable number of integron gene cassettes. The integron platform includes an integrase gene, ''intI1'' (a [[wikipedia:Site-specific_recombination|tyrosine site-specific recombinase]]), and a recombination site just upstream (''attI'') at which integron cassettes can be integrated. The cassettes are integrated in an orientation which allows their expression from a promoter, Pc, located within the integrase gene itself ([[:File:Fig-Tn402.1.png|Fig. Tn402.1]]) <ref name=":10">{{#pmid:26104695}}</ref><ref>{{#pmid:16845431}}</ref>. This definition is restrictive since it does not include the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] ends of the transposition enzymes. It raises the important question of how transposable elements such as [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] and a second transposon family, [[Transposons families/Tn7 family|Tn''7'']], members have acquired these gene cassette-acquiring systems. | ||
Tn''402'' ([https://www.ncbi.nlm.nih.gov/nuccore/U67194.4/ U67194.4]) is similar if not identical to Tn''5090'' ([https://www.ncbi.nlm.nih.gov/nuccore/AM993098 AM993098]) also isolated from R751<ref name=":6"> | [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] ([https://www.ncbi.nlm.nih.gov/nuccore/U67194.4/ U67194.4]) is similar if not identical to [https://tncentral.ncc.unesp.br/report/te/Tn5090-AM993098 Tn''5090''] ([https://www.ncbi.nlm.nih.gov/nuccore/AM993098 AM993098]) also isolated from R751<ref name=":6">{{#pmid:PMC205496}}</ref>. Both had subsequently been grouped into a family called [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] <ref name=":8">{{#pmid:8594337}}</ref> although we have elected here to call the family the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family based on its founding transposon. | ||
More recently, a second group of related transposons, the [[Transposons families/Tn402 family#The TnPfu1 group|Tn''Pfu1'' group]], was identified which include another integrase gene, ''IntI3'', oriented in the opposite direction to that in the [[Transposons families/Tn402 family#The Tn402 group and transposon decay.|Tn''402'' group]] <ref> | More recently, a second group of related transposons, the [[Transposons families/Tn402 family#The TnPfu1 group|Tn''Pfu1'' group]], was identified which include another integrase gene, ''IntI3'', oriented in the opposite direction to that in the [[Transposons families/Tn402 family#The Tn402 group and transposon decay.|Tn''402'' group]] <ref name=":17">{{#pmid:PMC6105817}}</ref>, and therefore acquired by a [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family independently of the type 1 integron, while a third group, [[Transposons families/Tn402 family#The Tn5053 group|Tn''5053'']], include genes for resistance to mercury salts instead of the integron platform <ref name=":0">{{#pmid:8387603}}</ref>.<br />[[File:Fig-Tn402.1.png|center|thumb|720x720px|'''Fig-Tn402.1.''' '''Organization of Tn''402''.''' [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] (Genbank accession number: [https://www.ncbi.nlm.nih.gov/nuccore/U67194.4/ U67194.4]) is shown as a pale yellow box. Open reading frames are shown as horizontal boxes where the arrowheads indicate the direction of translation: red, antibiotic resistance genes; lavender, transposase related genes (note that the start codon of TniA may be located further downstream than initially proposed as shown by the dotted outline following comparison with other members of the family – see text); purple, other; blue IntI; green boxes, integron cassette recombination sites and ''res'' sites; the terminal '''IRs''' (IRt, transposase proximal; IRi, integrase proximal) are also shown together with the short repeated sub-sites. These are shown in more detail below together with their relative orientations (gray arrows), and nucleotide sequences. The ''res'' site together with its six sub-repeats is shown in green. The figure was generated using [https://www.snapgene.com/ SnapGene].|alt=]] | ||
== Distribution == | |||
[https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family transposons are widely distributed in clinical and environmental isolates. A number of community studies have been carried out including from aquatic environments <ref name=":1">Jobling MG, Peters SE, Ritchie DA. Plasmid-borne mercury resistance in aquatic bacteria. FEMS Microbiol Lett. 1988;49:31–37.</ref>, permafrost <ref name=":2">{{#pmid:16084067}}</ref><ref name=":12">{{#pmid:21262357}}</ref><ref>{{#pmid:PMC1287632}}</ref><ref name=":21">{{#pmid:9159519}}</ref>, the [[wikipedia:Rhizosphere|rhizosphere]] <ref name=":3">{{#pmid:PMC7728920}}</ref><ref name=":4">{{#pmid:PMC97592}}</ref> and river and sewage sediments <ref name=":13">{{#pmid:PMC1540074}}</ref><ref name=":5">{{#pmid:PMC1636140}}</ref>. Because of their importance in environmental pollution, there has been much attention given to transposons carrying mercury resistance genes (see <ref>{{#pmid:25418738}}</ref>). These have been identified, for example, as part of plasmids isolated from the [[wikipedia:River_Mersey|river Mersey]] (U.K. <ref name=":1" />) in 1988 where an example was shown to share the same strict target specificity ([[Transposons families/Tn402 family#Insertion Sites: res Hunters.|Insertion Sites: ''res'' Hunters]]) and carry similar mercury resistance genes <ref name=":22">Hobman J, Kholodii G, Nikiforov V, Ritchie DA, Strike P, Yurieva O. The sequence of the mer operon of pMER327/419 and transposon ends of pMER327/419, 330 and 05. Gene. 1994 Aug;146(1):73–78.</ref> to [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''], isolated from a Russian mercury mine <ref name=":0" /> in 1984. | |||
A derivative, [https://tncentral.ncc.unesp.br/report/te/Tn50580-AM048832 Tn''50580''] carrying a complex set of mercury resistance genes was also identified in samples taken from sediment of the [[wikipedia:Nura_(river)|Nura]], a highly contaminated river in Kazakhstan <ref name=":5" />. Other examples include, related transposons and more complex [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] derivatives in permafrost samples many thousands of years old <ref name=":2" />; variants or transposon fragments with identical restriction maps (or partial sequences) in pre-antibiotic era strains in the Murray collection <ref>{{#pmid:PMC149298}}</ref> (''[[wikipedia:Enterobacteriaceae|Enterobacteriaceae]]'' isolated between 1917 and 1954 <ref>{{#pmid:6316165}}</ref>); association with plasmids also involved with degradation of the herbicide [[wikipedia:Atrazine|atrazine]] <ref>{{#pmid:PMC95461}}</ref>; and in a variety of bacteria including ''[[wikipedia:Pseudomonas_putida|Pseudomonas putida]]'', ''[[wikipedia:Escherichia_coli|Escherichia coli]]'', ''[[wikipedia:Enterobacter|Enterobacter]]'' and ''[[wikipedia:Klebsiella|Klebsiella]]'' from water and from soil from various geographical regions mainly, but not exclusively in Russia <ref>{{#pmid:11763242}}</ref>. | |||
Interestingly, [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']-like transposons and transposon fragments were identified in a survey of IncP group plasmids identified in the [[wikipedia:Rhizosphere|rhizosphere]] of greenhouse-grown plants <ref name=":3" />. Two included drug resistance gene cassettes but none were associated with mercury resistance. On the other hand, a study of an alfalfa [[wikipedia:Rhizosphere|rhizosphere]] in Germany revealed a number of plasmids carrying mercury resistance among which was pSN102 which carried the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']-family transposon [https://tncentral.ncc.unesp.br/report/te/Tn5718-AJ304453 Tn''5718''] <ref name=":4" /><ref name=":5" />. | |||
In view of the overuse of antibiotics, it is perhaps not surprising that [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] derivatives carrying antibiotic resistance integron cassettes have been found in sewage treatment plants (e.g. Tn''pRSB105'' carried by plasmid pRsB105;<ref name=":14">{{#pmid:PMC1828798}}</ref>) and also as rearranged fragments (e.g.<ref>{{#pmid:15817778}}</ref><ref>{{#pmid:19719593}}</ref><ref>{{#pmid:23212116}}</ref>) associated with other (rearranged TE) and also in estuarine waters <ref>{{#pmid:23831558}}</ref>. | |||
== Organization == | |||
=== Tn''402'' Terminal Repeated Sequences === | |||
Since it is now known that [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] and [https://tncentral.ncc.unesp.br/report/te/Tn5090-AM993098 Tn''5090''] are different isolates of the same element and given that [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] was the first described, we have elected to retain the name [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] throughout although it should be kept in mind that in some of the literature, the transposon is described as [https://tncentral.ncc.unesp.br/report/te/Tn5090-AM993098 Tn''5090'']. The nucleotide sequence of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] revealed a 7.4 kb region from R751 flanked by two 25bp IRs ([[:File:Fig-Tn402.1.png|Fig. Tn402.1]]). Two segments of Tn''402'' including each of the ends were found to be similar to the integron identified in the [[Transposons families/Tn3 family|Tn''3'' family]] transposon, [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21''] <ref>{{#pmid:PMC103744}}</ref>, and called Tn''5092'', and to a DNA segment embedded in [https://tncentral.ncc.unesp.br/report/te/Tn5086-CP054343 Tn''5086''] called Tn''5093'' <ref name=":6" /><ref>{{#pmid:PMC203974}}</ref>. | |||
In all cases, the '''IR''' were flanked by 5 bp direct target repeats different for each Tn. All three Tn included a type I integron integrase ''intI1'' at one end. The '''IRs''' are referred to as IRi for the copy proximal to the integrase end and IRt for the copy proximal to the opposite (transposase) end. The transposon ends also include several 19 bp repeats: t1, t2 and t3 located at the IRt end with t2 oriented inversely and i1, i2, i3 and i4 located at the IRi transposon end ([[:File:Fig-Tn402.1.png|Fig. Tn402.1]]). A fourth repeat, t4, can be identified by inspection. These are binding sites for the transposase <ref name=":9">{{#pmid:PMC29607}}</ref> and their differential organisation at each end would serve to distinguish between the ends in the transposition process as has been shown for [[Transposons families/Tn7 family|transposon Tn''7'']] <ref>Lichtenstein C, Brenner S. Site-Specific Properties of Tn7 Transposition into the E. coil Chromosome. Mol Gen Genet. 1981;183:380–387.</ref><ref>McKown RL, Waddell CS, Arciszewska LK, Craig NL. Identification of a transposon Tn7-dependent DNA-binding activity that recognizes the ends of Tn7. ProcNatlAcadSciUSA. 1987;84:7807–7811.</ref><ref>{{#pmid:PMC328805}}</ref> and [[wikipedia:Bacteriophage_Mu|bacteriophage Mu]] <ref>{{#pmid:6094016}}</ref><ref>{{#pmid:PMC3536463}}</ref> ([[:File:Fig-Tn402.2.png|Fig. Tn402.2]]) and members of the [[IS Families/IS21 family|IS''21'' family]] <ref>{{#pmid:PMC98933}}</ref>. | |||
<br /> | <br /> | ||
[[File:Fig-Tn402. | [[File:Fig-Tn402.2.png|center|thumb|600x600px|'''Fig-Tn402.2.''' '''Alignment of TniA and TnsB.''' The region carrying the catalytic DDE triad of TniA ([https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']) and TnsB ([https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7'']) are shown. They were generated using [[wikipedia:Clustal|Clustal]] and the figure was made using Jalview Using standard clustal coloring for amino acid residues.|alt=]] | ||
< | |||
==== | |||
Tn''402'' | [[File:Fig-Tn402.3.png|thumb|550x550px|'''Fig. Tn402.3.''' '''Multiple Repeats in Transposon Ends.''' The transposon ends are shown as yellow boxes with the short repeat sequences and their relative orientation indicated by blue arrows. Tn''402'' <ref name=":6" />; [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] (<ref>{{#pmid:PMC328805}}</ref> ; [[wikipedia:Bacteriophage_Mu|bacteriophage Mu]] <ref>{{#pmid:6094016}}</ref>.]] | ||
==== Tn''402'' Integron Cassettes ==== | |||
[https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] carries three integron cassettes. One of these contains the trimethoprim resistance gene'', dhf''rIIc (dihydrofolate reductase type IIc) originally identified in the first transposition experiments <ref name=":7" />. This is located proximal to the integron platform promoter ([[:File:Fig-Tn402.1.png|Fig. Tn402.1]]). Another, ''orfD'', of unkown function but observed in other integrons, is located downstream and this is followed by a gene encoding the exporter, ''[https://www.uniprot.org/uniprot/P0AGD0 qacE]''/[https://www.uniprot.org/uniprot/O87868 ''qaceH'']. | |||
==== Tn''402'' Transposition-Related Genes ==== | |||
Four transposition-related ''orfs'' were also identified. Three of these, ''tniA'', ''tniB'' and ''orf6,'' now called ''tniQ'', <ref name=":8" /> specify proteins of 559, 302 and 405 amino acids respectively. They occur in that order and are all oriented in the same direction ([[:File:Fig-Tn402.1.png|Fig. Tn402.1]]). They probably constitute an operon expressed from a predicted single promoter <ref name=":6" />. The fourth gene, initially called ''tniC'', but now renamed as ''tniR'' (see below) encodes a 207 amino acid protein. | |||
===== Tn''402'' Transposition-Related Genes: ''tniA'' ===== | |||
TniA was predicted to be a transposase since it shows some weak similarity to the transposon [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] transposase TnsB ([[:File:Fig-Tn402.2.png|Fig. Tn402.2]]), and to the Tn''552'' transposase (25% identity) and includes a DDE motif typical of the catalytic site of many transposases. ''tniA'' is located close to IRt. Transposon [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''], TnsB, which binds to the multiple short 22bp repeated sequences located in the left and right [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] ends ([[:File:Fig-Tn402.3.png|Fig. Tn402.3]]), suggesting that TniA may bind to the 19 bp repeated sequences in the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] ends. This has now been demonstrated using DNase and hydroxy-radical footprinting approaches <ref name=":9" />. It should be noted that there is some ambiguity in the position of the TniA initiation codon (see below; [[:File:Fig-Tn402.6.png|Fig. Tn402.6]]). | |||
===== Tn''402'' Transposition-Related Genes: ''tniB'' ===== | |||
The product of the second orf, TniB ([[:File:Fig-Tn402.1.png|Fig. Tn402.1]]), contains predicted nucleotide triphosphate binding sites and was proposed to be involved in nucleotide triphosphate hydrolysis <ref name=":6" />. It exhibits 21% identity to the [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] TnsC protein, an [[wikipedia:ATPase|ATPase]] <ref>{{#pmid:PMC1207843}}</ref> which binds non-specifically to DNA in the presence of ATP <ref>{{#pmid:PMC312388}}</ref>, and is required for transposition. TnsC also binds to the TnsB component the heterodimeric TnsA/TnsB transposase and couples it to a protein involved in targeting [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] insertion, TnsD <ref>{{#pmid:PMC4104917}}</ref>. TniB also shows 24% identity to the MuB protein, also a DNA-dependent [[wikipedia:ATPase|ATPase]] that preferentially stimulates intermolecular DNA strand transfer in [[wikipedia:Bacteriophage_Mu|bacteriophage Mu]] <ref>{{#pmid:PMC304283}}</ref>. | |||
===== Tn''402'' Transposition-Related Genes: ''tniQ'' ===== | |||
The third gene ([[:File:Fig-Tn402.1.png|Fig. Tn402.1]]) was called orf6 <ref name=":6" /> but has since been named ''tniQ'' <ref name=":8" /> and its product did not exhibit similarity to known proteins <ref name=":6" />. It is positioned such that its initiation codon overlaps the termination codon of the upstream ''tniB'' gene (see below; [[:File:Fig-Tn402.7.png|Fig. Tn402.7]]) indicating that expression of the two gene products is translationally coupled. | |||
===== Tn''402'' Transposition-Related Genes: ''tniR'' ===== | |||
The product of the fourth gene belongs to the invertase/resolvase family of [[wikipedia:Site-specific_recombination|site-specific recombinases]]. It has significant identity (67%) to the resolvase of the [[Transposons families/Tn3 family|Tn''3'' family transposon]], [https://tncentral.ncc.unesp.br/report/te/Tn5393-M95402.1 Tn''5393'']'','' 47% identity to the Gin recombinase of [[wikipedia:Bacteriophage_Mu|bacteriophage Mu]] <ref>Plasterk RH, Brinkman A, van de Putte P. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. ProcNatlAcadSciUSA. 1983;80:5355–5358.</ref>. For this reason, it is now generally referred to as ''tniR'' for Resolvase. It is probably expressed separately from the other three genes and appears to have its own promoter <ref name=":6" /> (see below; [[:File:Fig-Tn402.5a.png|Fig. Tn402.5]] '''A'''). | |||
==== Tn''5053'' Terminal Repeated Sequences ==== | |||
More information was obtained from the related [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] ([https://www.ncbi.nlm.nih.gov/nuccore/L40585.1/ L40585.1])'','' a mercury resistance transposon isolated from the chromosome of a mercury resistant ''[[wikipedia:Xanthomonas|Xanthomonad]]'' from a mercury mine <ref name=":0" />. [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] carries a set of transposition genes which are very similar to those of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']/[https://tncentral.ncc.unesp.br/report/te/Tn5090-AM993098 Tn''5090'']. Like other members of the family it is bordered by 25 bp inverted repeats ([[:File:Fig-Tn402.4.png|Fig. Tn402.4]]) and includes short ~19 bp repeat sequences at its ends. | |||
= | Careful inspection of the [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] sequence identifies additional 19bp repeat sequences than those reported <ref name=":0" />, as in the case of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']/[https://tncentral.ncc.unesp.br/report/te/Tn5090-AM993098 Tn''5090''] ([[:File:Fig-Tn402.1.png|Fig. Tn402.1]]). In this case there are 4 copies at the IRt end and three at the equivalent of the IRi end. Also, in the original article, it appears that those at the IRt and IRi ends have been exchanged compared to [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']/[https://tncentral.ncc.unesp.br/report/te/Tn5090-AM993098 Tn''5090'']. [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] carries a full complement of transposition genes ''tniA,B,Q'' and ''R'' but, instead of the integron recombination platform and antibiotic resistance gene cassettes, the transposon carries a mercury resistance operon ([[:File:Fig-Tn402.4.png|Fig. Tn402.4]]). | ||
[[File:Fig-Tn402.4.png|center|thumb|640x640px|'''Fig. Tn402.4.''' '''Organization of Tn''5053''.''' [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] (Genbank accession number: [https://www.ncbi.nlm.nih.gov/nuccore/L40585.1/ L40585.1]) is shown as a pale yellow box. Open reading frames are shown as horizontal boxes where the arrowheads indicate the direction of translation: lavender, transposase related genes (note that the N-terminal end of TniA is ambiguous – see text); yellow, mercury resistance genes; green boxes, res sites; the terminal IRs (IRt, transposase proximal; IRi, transposase distal) are also shown together with the short repeated sub-sites. The res site together with its six sub-repeats is shown in green. Irt and IRi are shown in more detail below together with their relative orientations (grey arrows), and nucleotide sequences. The sequence at the bottom of the figure indicates the potential promoter (-35 and -10 elements in red) for tniR expression. The figure was generated using [https://www.snapgene.com/ SnapGene].]] | |||
== | == Tn''5053'' Functional analysis of Transposition-Related Genes == | ||
Transposition of a collection of insertion and deletion mutants of [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] introduced into ''tniA'', ''tniB'' and ''tniQ'' was tested in a conjugation (mating out <ref name=":34">Galas DJ, Chandler M. Structure and stability of Tn9-mediated cointegrates. J Mol Biol. 1982 Jan;154(2):245–272.</ref>) assay using an RP4 plasmid derivative as a target <ref name=":8" />. Transposition was undetectable in all mutants but could be complemented by supplying a set of [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] genes ''in trans'' from a third compatible plasmid. | |||
It was also shown that the set of three genes from [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] were capable of complementing the mutant [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] derivatives <ref name=":8" />. To address whether the mutations may have a polar effect on the downstream genes, mutations in the complementing set of genes were used: a [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] ''tniA'' mutant could be complemented by a ''tniA'' operon with a wildtype ''tniA'' and carrying mutations in ''tniB'' and ''tniQ'' , while a ''tniB'' mutant could be complemented by a ''tniQ''/''tniR'' mutant. It is unclear whether a ''tniB'' mutant could be complemented by a ''tniA/tniQ/tniR'' mutant raising the formal possibility that some polarity effects may occur. | |||
[[File:Fig-Tn402.5a.png|center|thumb|600x600px|'''Fig. Tn402.5.''' '''Organization of [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] and [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] res sites.''' '''A)''' Tn''402'' (Genbank accession number: [https://www.ncbi.nlm.nih.gov/nuccore/U67194.4 U67194.4]) is shown as a pale yellow box. TniQ and TniR open reading frames are shown as horizontal lavender boxes, where the arrowheads indicate the direction of translation. The res site and subsites are shown as green box and green arrows respectively. ]] | |||
<br /> | <br /> | ||
[[File:Fig-Tn402. | [[File:Fig-Tn402.5b.png|center|thumb|600x600px|'''Fig. Tn402.5.''' '''Organization of [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] and [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] res sites.''' '''B)''' DNA sequence of the intergenic region showing th''e tn''iQ and ''tniR'' sequences in lavender, and the repeated sequences as green arrows. The dinucleotide at which recombination is proposed to take place between r1 and r2 is shown in red. -35 and -10 sequences for ''tniR'' are underlined in red. The sequences below represent the base changes observed in the Tn402 sequence. This suggests that the correct translation start codon may be located somewhat downstream from that proposed for [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] since the ATG originally proposed does not occur in Tn''402''.]] | ||
<br /> | <br /> | ||
== Tn''5053'' Functional Analysis of TniR and the ''res'' site == | |||
Tn'' | As might be expected from the similarity of ''tniR'' to site-specific resolvases, [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] appears to transpose via a cointegrate intermediate. Ablation of the ''tniR'' gene and part of the tniQ gene resulted in a transposon unable to undergo transposition, presumably because of the defective TniQ. Complementation using the entire set of transposition genes resulted in high transposition levels. In the absence of TniR in the complementing plasmid, cointegrates carrying both donor and recipient plasmid antibiotic resistance markers were identified. Resolution required the presence of only TniR and not TniA, B or Q. | ||
The recombination site was located using a system with cloned Tn restriction fragments from a region upstream of ''tniR'' and the “kinetics“ of resolution was followed. It was concluded that a region close to the 5’ end of the ''tniR'' gene which, on its own resulted in slow resolution, represented the principal resolution site while a region stretching upstream into the 3’ end of the ''tniQ'' gene was required for robust activity and therefore presumably carried secondary stimulating sequences. This region includes six 14 bp repeat sequences with the consensus PyTGTCACPuT(NT)(NT)NC(C/G), labelled r1-r6 (shown in detail in [[:File:Fig-Tn402.5a.png|Fig. Tn402.5]] '''A''' and [[:File:Fig-Tn402.5b.png|Fig. Tn402.5]] '''B'''). | |||
Most site-specific resolution systems include multiple short repeated sequences which serve as recombinase binding sites permitting the assembly of architecturally precise nucleoprotein complexes to tightly control recombination. The repeats include r1 and r2 which are inverted, almost abut the start of the ''tniR'' gene and are separated by a dinucleotide, AA, proposed as the crossover point in the resolution recombination reaction ([[:File:Fig-Tn402.4.png|Fig. Tn402.4]]). Additional copies are observed further upstream and located within the 3’ end of ''tniQ'' ([[:File:Fig-Tn402.5a.png|Fig. Tn402.5]] '''A''' and [[:File:Fig-Tn402.5b.png|Fig. Tn402.5]] '''B''') suggesting that TniQ expression and resolution are intimately coupled. Further analysis is necessary to provide direct evidence that these repeated sequences are TniR binding sites and to establish a detailed recombination mechanism. | |||
====== | === TniR Expression and Translation Initiation === | ||
A potential promoter <ref name=":8" /> for ''tniR'' expression is also located in this region ([[:File:Fig-Tn402.5b.png|Fig. Tn402.5]] '''B''') and is clearly well placed to be regulated by TniR binding. [[:File:Fig-Tn402.5b.png|Fig. Tn402.5]] '''B''' also shows that the equivalent region of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']/[https://tncentral.ncc.unesp.br/report/te/Tn5090-AM993098 Tn''5090''] is very similar with related repeat sequences and potential promoter elements. However, there is additional G residue and changes at the potential TniR ATG codon compared to those observed in [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053'']. However, there is a suitably placed in phase '''ATG codon''' slightly further downstream in both [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']/[https://tncentral.ncc.unesp.br/report/te/Tn5090-AM993098 Tn''5090''] and in [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] which suggests that this maybe the true initiation codon for TniR. | |||
======Tn''402'' | === TniA Expression and Translation Initiation === | ||
In addition to the ambiguity of the TniR initiation codon, a similar question arises concerning identification of the initiation codon of TniA ([[:File:Fig-Tn402.6.png|Fig. Tn402.6]]). Certain family members (e.g. [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] and integron members) have an ''orf'' with an initiation codon upstream of the final t repeat ([[:File:Fig-Tn402.6.png|Fig. Tn402.6]]) whereas others (e.g. [https://tncentral.ncc.unesp.br/report/te/Tn502-EU306743 Tn''502''] and [https://tncentral.ncc.unesp.br/report/te/Tn512-EU306744 Tn''512'']) appear to initiate further downstream. | |||
=== | Alignment of the DNA sequences shows that all members are very similar in sequence in this region but that those with the downstream start codon each contain a 2 bp insertion which places the upstream region out of phase ([[:File:Fig-Tn402.6.png|Fig. Tn402.6]]). Since [https://tncentral.ncc.unesp.br/report/te/Tn502-EU306743 Tn''502''] and [https://tncentral.ncc.unesp.br/report/te/Tn512-EU306744 Tn''512''] and [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] are fully functional <ref name=":0" /><ref name=":23">{{#pmid:PMC247445}}</ref><ref name=":24">{{#pmid:PMC2838034}}</ref>, it seems possible that it is the downstream start codon that is used in all family members ([https://tncentral.ncc.unesp.br/report/te/Tn402.10-CP040126 Tn''402.9''] TniA). But this will need experimental confirmation.[[File:Fig-Tn402.6.png|center|thumb|640x640px|'''Fig. Tn402.6.''' '''The Tn''402'' Family TniA Initiation Codon.''' The Figure shows DNA alignment of selected [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family transposons against the 5’ end of ''tniA'' from [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']. Both [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] DNA strands are shown at the top of the figure. [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] is represented by a pale yellow box. The ''tniA'' orf is shown as a horizontal lavender box. The region encircled by a dotted black line is the region which is absent in the [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] group. The sequences of selected [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] members are aligned below. The transposon together with the sequence accession number from where each transposon was extracted are shown on the left. All transposons carry the ATG codon (boxed in green) proposed to be the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] TniA initiation codon. However, four transposons ([https://tncentral.ncc.unesp.br/report/te/Tn502-EU306743 Tn''502'']-[https://www.ncbi.nlm.nih.gov/nuccore/EU306743 EU306743]; [https://tncentral.ncc.unesp.br/report/te/Tn512-EU306744 Tn''512'']-[https://www.ncbi.nlm.nih.gov/nuccore/EU306744 EU306744]; [https://tncentral.ncc.unesp.br/report/te/Tn5058-Y17897 Tn''5058'']-[https://www.ncbi.nlm.nih.gov/nuccore/Y17897 Y17897]; and [https://tncentral.ncc.unesp.br/report/te/Tn6007-EU591509.1 Tn''6007'']-[https://www.ncbi.nlm.nih.gov/nuccore/EU591509.1 EU591509.1]) carry a 2 bp insertion, TG, which forces the resulting protein out of frame. Moreover, another two transposons ([https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053'']-[https://www.ncbi.nlm.nih.gov/nuccore/L40585.1 L40585.1] and In_TnCfr-[https://www.ncbi.nlm.nih.gov/nuccore/CP016763.1 CP016763.1]) carry instead, a 2 bp insertion, TT, one base pair upstream which also forces the resulting protein out of frame. However, the TniA initiation codon proposed for all six transposons (ATG, boxed in green) is also present in [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']-[https://www.ncbi.nlm.nih.gov/nuccore/U67194.4 U67194.4] and in the integrons [https://tncentral.ncc.unesp.br/report/te/In2-AF071413 In2]-[https://www.ncbi.nlm.nih.gov/nuccore/AF071413 AF071413] and In73-[https://www.ncbi.nlm.nih.gov/nuccore/KY978631 KY978631] as well as in all other known [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family members. The most parsimonious explanation is that this ATG is the ''tniA'' initation codon for all family members. The figure was generated using [https://www.snapgene.com/ SnapGene].]] | ||
The | |||
== | == TniB Expression, Translation Initiation and Translational Coupling with TniA == | ||
A similar ambiguity arises with TniB. A DNA alignment ([[:File:Fig-Tn402.7.png|Fig. Tn402.7]] '''A'''), however, not only shows a high degree of identity but suggests that there may be some form of translational coupling in the expression of TniA and TniB. There are potential TniB GTG start codons 1bp upstream and 2bp downstream of the predicted TniA termination codon (TAG) <ref name=":11">{{#pmid:PMC6728339}}</ref>. | |||
Although we have chosen to use the downstream GTG ([[:File:Fig-Tn402.9a.png|Fig. Tn402.9]] TniB), it should be remembered that if the upstream GTG is used, TniB would include an N-terminal extension of three amino acids, MVA. Note that the 2 mutations shown in the figure are synonymous (ATT/ATC) and conservative (GAC/GAA) respectively. | |||
== TniQ Expression, Translation Initiation and Translational Coupling with TniB == | |||
For many examples of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family transposons, the initation codon of TniQ (GTG) overlaps the termination codon (TGA) of TniB ([[:File:Fig-Tn402.7.png|Fig. Tn402.7]] '''B'''), a situation typical of translational coupling <ref name=":11" />. All members of the family so far identified show high conservation and all contain the overlapping GTGA initiation/termination codons. Although some family members had included annotated TniB and or TniQ genes which had internal start codons, the most parsimonious interpretation is that the entire family follow identical initiation and termination rules ([[:File:Fig-Tn402.9a.png|Fig. Tn402.9]] TniQ). | |||
<br /> | |||
[[File:Fig-Tn402.7.png|center|thumb|640x640px|'''Fig. Tn402.7.''' '''TniA-B and TniB-Q Junctions:''' T'''ranslational Coupling?''' The figure presents an alignment of the DNA of selected transposons at the junction between ''tniA'' and ''tniB'' ('''A''') and between ''tniB'' and ''tniQ'' ('''B'''). Both [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] DNA strands are shown at the top of the figure. [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] is represented by a pale yellow box. The ''tni'' ''orfs'' are shown as horizontal lavender boxes. Termination codons are boxed in red and initiation codons in green. '''A)''' All family members include a TAG TniA stop codon preceded 1bp upstream and followed, 2 bp downstream by potential TniB GTG initiation codons. '''B)''' All family members carry overlapping (GTGA) TniQ initiation codons GTG and TniB termination codons TGA. Although some family members had included annotated TniB and or TniQ genes which had internal start codons, the most parsimonious interpretation is that the entire family follows identical initiation and termination rules. Note that certain integrons are truncated at the 3’ end of the ''tniB'' ''orf'' due to IS insertions ([https://tncentral.ncc.unesp.br/report/te/IS1326_IS1353-AF071413 IS''1326''::IS''1353''] in In2-[https://www.ncbi.nlm.nih.gov/nuccore/AF071413 AF071413] and in its deletion in [https://tncentral.ncc.unesp.br/report/te/In_TnAs3-CP000645.1 In_Tn''As3'']-[https://www.ncbi.nlm.nih.gov/nuccore/CP000645.1 CP000645.1] and [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017'']-[https://www.ncbi.nlm.nih.gov/nuccore/NC_022242.1 NC_022242.1]. The figure was generated using [https://www.snapgene.com/ SnapGene].]] | |||
== | == Diversity == | ||
There are three principal groups which constitute the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family based on the type of passenger genes they carry. The first, [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''], includes those which have acquired an integron recombination platform of type 1 with an integrase IntI1 derivative. The second, [https://tncentral.ncc.unesp.br/report/te/TnPfu1-LC331665.1 Tn''Pfu1''], is a small group composed of members which have integrated a type 3 integron recombination platform with an IntI3 integrase. The third group, [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''], includes members which do not carry integron function but instead carry mercury resistance genes. | |||
A phylogenetic tree of the concatenated TniA,B,Q and R proteins ([[:File:Fig-Tn402.8.png|Fig. Tn402.8]]) indicated that [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] group members carrying mercury resistance genes tend to be clustered with [https://tncentral.ncc.unesp.br/report/te/Tn502-EU306743 Tn''502''] and [https://tncentral.ncc.unesp.br/report/te/Tn512-EU306744 Tn''512''] although several are intermingled with [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] IntI1 carrying transposons. This is perhaps not surprising since many [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] members were selected using a BLAST search. A similar argument can be proposed for the three [https://tncentral.ncc.unesp.br/report/te/TnPfu1-LC331665.1 Tn''Pfu1''] related transposons which carry an IntI3 gene. | |||
[[File:Fig-Tn402.8.png|center|thumb|800x800px|'''Fig. Tn402.8.''' '''Phylogeny of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] Family Transposition Enzymes.''' TnpA protein sequences retrieved from the TnCentral curated data set were aligned with MAFFT 7.309, and their best-fit evolutionary models were predicted with ProTest 3.2.4. A maximum-likelihood tree was reconstructed with RaxML 8.2.9 using a bootstrap value of 1,000. The final tree was visualized in [http://tree.bio.ed.ac.uk/software/figtree FigTree 1.4.4] and edited with [http://www.inkscape.org Inkscape 0.92.4]. The colored circles indicate which group the transposons belong to. The insert gives the color code for members of family members with IntI1, InI3 and those carrying mercury resistance genes. Tn included together with the accession numbers from which they were drawn are: Tn''402''-[https://www.ncbi.nlm.nih.gov/nuccore/U67194.4 U67194.4;] Tn''402.1''-[https://www.ncbi.nlm.nih.gov/nuccore/GQ857074.1 GQ857074.1]; Tn''402.2''-[https://www.ncbi.nlm.nih.gov/nuccore/CP045551 CP045551]; Tn''402.3''-[https://www.ncbi.nlm.nih.gov/nuccore/CP063457 CP063457]; Tn''402.4''-[https://www.ncbi.nlm.nih.gov/nuccore/MW150990 MW150990]; Tn''402.5''-[https://www.ncbi.nlm.nih.gov/nuccore/MH392234 MH392234]; Tn''402.6''-[https://www.ncbi.nlm.nih.gov/nuccore/KT345947 KT345947]; Tn''402.8''-[https://www.ncbi.nlm.nih.gov/nuccore/MF490433 MF490433]; Tn''402.10''-[https://www.ncbi.nlm.nih.gov/nuccore/CP040126 CP040126]; Tn''502''-[https://www.ncbi.nlm.nih.gov/nuccore/EU306743 EU306743]; Tn''512''-[https://www.ncbi.nlm.nih.gov/nuccore/EU306744 EU306744]; In_TnAs3-[https://www.ncbi.nlm.nih.gov/nuccore/CP000645.1 CP000645.1]; In31-[https://www.ncbi.nlm.nih.gov/nuccore/AJ223604 AJ223604]; In32-[https://www.ncbi.nlm.nih.gov/nuccore/L36547 L36547]; In34-[https://www.ncbi.nlm.nih.gov/nuccore/AY123253 AY123253]; In73-[https://www.ncbi.nlm.nih.gov/nuccore/KY978631 KY978631]; In_Tn1721.1-[https://www.ncbi.nlm.nih.gov/nuccore/HQ730118.1 HQ730118.1]; In0-[https://www.ncbi.nlm.nih.gov/nuccore/U49101 U49101]; In2-[https://www.ncbi.nlm.nih.gov/nuccore/AF071413 AF071413]; Tn''6112''-[https://www.ncbi.nlm.nih.gov/nuccore/HQ423158.1 HQ423158.1]; TnpRSB105-[https://www.ncbi.nlm.nih.gov/nuccore/DQ839391.1 DQ839391.1]; Tn''5053''-[https://www.ncbi.nlm.nih.gov/nuccore/L40585.1 L40585.1]; Tn''5053.1''-[https://www.ncbi.nlm.nih.gov/nuccore/CP002451 CP002451]; Tn''5053.2''-[https://www.ncbi.nlm.nih.gov/nuccore/CP024682 CP024682]; Tn''5053.3''-[https://www.ncbi.nlm.nih.gov/nuccore/GQ983559 GQ983559]; Tn''5053.4''-[https://www.ncbi.nlm.nih.gov/nuccore/CP001919 CP001919]; Tn''5053.6''-[https://www.ncbi.nlm.nih.gov/nuccore/JX469833 JX469833]; Tn''5053.7''-[https://www.ncbi.nlm.nih.gov/nuccore/AM261760 AM261760]; Tn''5053.8''-[https://www.ncbi.nlm.nih.gov/nuccore/CP017991 CP017991]; Tn''5053.9''-[https://www.ncbi.nlm.nih.gov/nuccore/CP048650 CP048650]; Tn''6008''-[https://www.ncbi.nlm.nih.gov/nuccore/EU591509.1 EU591509.1]; In_TnCfrpOZ172-[https://www.ncbi.nlm.nih.gov/nuccore/CP016763.1 CP016763.1]; Tn''6007''-[https://www.ncbi.nlm.nih.gov/nuccore/EU591509.1 EU591509.1]; Tn''5718''-[https://www.ncbi.nlm.nih.gov/nuccore/AJ304453 AJ304453]; Tn''5058''-[https://www.ncbi.nlm.nih.gov/nuccore/Y17897 Y17897]; Tn''Pfu1''-[https://www.ncbi.nlm.nih.gov/nuccore/LC331665.1 LC331665.1]; Tn''Pfu1.1''-[https://www.ncbi.nlm.nih.gov/nuccore/LC589064 LC589064]; Tn''Pfu1.2''-[https://www.ncbi.nlm.nih.gov/nuccore/LC589062 LC589062]; Tn''7017''-[https://www.ncbi.nlm.nih.gov/nuccore/NC_022242.1 NC_022242.1]; TnpHS87a-[https://www.ncbi.nlm.nih.gov/nuccore/KR106190.1 KR106190.1]; TnXorYNA12-[https://www.ncbi.nlm.nih.gov/nuccore/HQ662557.1 HQ662557.1]; TnpTB11-[https://www.ncbi.nlm.nih.gov/nuccore/AJ744860 AJ744860].]] | |||
When considered individually, the different gene products are very closely related: TniA ([[:File:Fig-Tn402.9a.png|Fig. Tn402.9]] see the slide show below); TniB ([[:File:Fig-Tn402.9a.png|Fig. Tn402.9]] see the slide show below); TniQ ([[:File:Fig-Tn402.9a.png|Fig. Tn402.9]] see the slide show below) and TniR ([[:File:Fig-Tn402.9a.png|Fig. Tn402.9]] see the slide show below). | |||
The fact that the TniA,B,Q and R proteins are highly conserved ([[:File:Fig-Tn402.9a.png|Fig. Tn402.9]]) whereas the DNA sequences are more variable (see [[:File:Fig-Tn402.11.png|Fig. Tn402.11]] and [[:File:Fig-Tn402.13.png|Fig. Tn402.13]]) (and therefore the DNA variability mainly introduces synonymous mutations together with some conservative amino acid changes; [[:File:Fig-Tn402.9a.png|Fig. Tn402.9]]) suggests either that the phylogenetic branches are quite recent or that there is significant selective pressure on the protein ensemble. | |||
= | The major differences between each of the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family members lie in the passenger genes which they carry.<gallery mode="slideshow"> | ||
File:Fig-Tn402.9a.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9b.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9c.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9d.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9e.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9f.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9g.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9h.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9i.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9j.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9k.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9l.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9m.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9n.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9o.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
File:Fig-Tn402.9p.png|'''Fig. Tn402.9.''' Global alignments of TniA,B,Q and R proteins from a collection of Tn''402'' family members. | |||
</gallery> | |||
== | == The Tn''402'' group and transposon decay == | ||
[[File:Fig-Tn402.10.png|thumb|640x640px|'''Fig. Tn402.10.''' Comparison of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family members showing the “decay” in the TniABQR genes. [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] is shown as a pale yellow box. Open reading frames are shown as horizontal boxes where the arrowheads indicate the direction of translation. Lavender, transposase related genes; red, antibiotic resistance genes; purple, other; blue, IntI ; the terminal IRs (IRt, transposase proximal; IRi, transposase distal) are also shown together with the short repeated sub-sites. The res site together with its six sub-repeats is shown in green, as are the attI and attC recombination sites involved in acquiring integron cassettes. Above is shown the alignment of various [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] members (names listed on the right). The extent of homology with different parts of Tn''402'' are shown as red horizontal lines. Small triangles indicate insertions relative to [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']. The figure was generated using [https://www.snapgene.com/ SnapGene].]] | |||
As stated above, there has been some ambiguity in the literature concerning the nomenclature of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family members as transposons or as integrons and that this has been due to decay in the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] tniABQR transposition module. In this group, the antibiotic resistance genes are oriented towards and downstream of the transposition genes ([[:File:Fig-Tn402.1.png|Fig. Tn402.1]]). [[:File:Fig-Tn402.10.png|Fig. Tn402.10]] shows an alignment of an illustrative group of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] elements. | |||
All the examples include the right (integrase proximal) [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] end, IRi and its 19 bp repeated sequences together with the integron integrase, ''intI1'' gene, the binding site for the host [[wikipedia:Repressor_lexA|LexA]] protein which couples IntI1 expression to the host [[wikipedia:SOS_response|SOS system]] <ref name=":10" /><ref>{{#pmid:PMC2958807}}</ref><ref>{{#pmid:20707672}}</ref>, the integron cassette promoter P<sub>c</sub> which drives expression of the genes carried in the integron cassettes, and the ''attI'' recombination site which serves in the acquisition of integrons cassettes <ref name=":10" />. | |||
This is followed by the integron cassettes which vary from element to element and are indicated by the small arrow heads. [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] ([https://www.ncbi.nlm.nih.gov/nuccore/U67194.4 U67194.4]) itself carries three cassettes: ''[https://card.mcmaster.ca/ontology/40527 qacH]''; ''orfD'', a gene of unknown function; and the dihydrofolate reductase gene, ''[https://card.mcmaster.ca/ontology/39456 dfrB3]''. The [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 IS''402''] family member with the lowest number of integron cassettes is [https://tncentral.ncc.unesp.br/report/te/In_Tn1721.1-HQ730118.1 In_Tn''1721.1''] ([https://www.ncbi.nlm.nih.gov/nuccore/HQ730118.1 HQ730118.1]) which includes only ''[https://card.mcmaster.ca/ontology/40527 qacH]'' and its recombination sites while [https://tncentral.ncc.unesp.br/report/te/Tn402.5-MH392234 Tn''402.5''] ([https://www.ncbi.nlm.nih.gov/nuccore/MH392234 MH392234]) <ref name=":3" /> carries, in addition, a second cassette with a gene of unknown function. | |||
Within this group, [https://tncentral.ncc.unesp.br/report/te/Tn402.5-MH392234 Tn''402.5''] ([https://www.ncbi.nlm.nih.gov/nuccore/MH392234 MH392234]), [https://tncentral.ncc.unesp.br/report/te/In_Tn1721.1-HQ730118.1 In_''Tn1721.1''], [https://tncentral.ncc.unesp.br/report/te/Tn402.4-MW150990 Tn''402.4''] ([https://www.ncbi.nlm.nih.gov/nuccore/MW150990 MW150990]), [https://tncentral.ncc.unesp.br/report/te/Tn6112-HQ423158.1 Tn''6112''],([https://www.ncbi.nlm.nih.gov/nuccore/HQ423158 HQ423158] ) <ref>{{#pmid:PMC3019745}}</ref>, [https://tncentral.ncc.unesp.br/report/te/TnpHS87a-KR106190.1 Tn''pHS87''a] ([https://www.ncbi.nlm.nih.gov/nuccore/KR106190 KR106190]) <ref>{{#pmid:PMC4739549}}</ref>, and [https://tncentral.ncc.unesp.br/report/te/TnXorYNA12-HQ662557.1 Tn''XorYNA12''] ([https://www.ncbi.nlm.nih.gov/nuccore/HQ662557 HQ662557]) <ref>{{#pmid:PMC3578876}}</ref> all carry a full complement of the transposition module and a variety of different integron cassettes. | |||
A second group has lost both ''tniR'' and ''tniQ'' genes. These include: [https://tncentral.ncc.unesp.br/report/te/In0-U49101 In0] ([https://www.ncbi.nlm.nih.gov/nuccore/U49101 U49101]) <ref>{{#pmid:PMC206418}}</ref> where they, together with the 3’ end of ''tniB'', have been replaced by an insertion of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1326 IS''1326''] upstream of a GNAT cassette; [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''] ([https://www.ncbi.nlm.nih.gov/nuccore/NC_022242.1 NC_022242.1]) <ref>{{#pmid:23928025}}</ref> in which the [https://tncentral.ncc.unesp.br/report/te/In0-U49101 In0] GNAT cassette abuts a further 3’ deleted ''tniB'' (presumably following excision of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1326 IS''1326''] copy); [https://tncentral.ncc.unesp.br/report/te/TnpRSB105-DQ839391.1 Tn''pRSB105''] ([https://www.ncbi.nlm.nih.gov/nuccore/DQ839391 DQ839391]) <ref name=":14" /> which has a similar tniB-GNAT junction but in which a tip of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1326 IS''1326''] (GGCCTG) has been retained; [https://tncentral.ncc.unesp.br/report/te/In2-AF071413 In2] ([https://www.ncbi.nlm.nih.gov/nuccore/AF071413 AF071413]) which contains an insertion of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1353 IS''1353''] into the ''istA'' gene of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1326 IS''1326''], a common feature of a number of integron structures; [https://tncentral.ncc.unesp.br/report/te/In_Tn4-KY749247.1 In_Tn''4''] ([https://www.ncbi.nlm.nih.gov/nuccore/KY749247.1 KY749247.1]) <ref>{{#pmid:28359666}}</ref> which, except for the absence of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1353 IS''1353''] insertion, is identical to [https://tncentral.ncc.unesp.br/report/te/In2-AF071413 In2]; [https://tncentral.ncc.unesp.br/report/te/In_TnAs3-CP000645.1 In_Tn''As3''] ([https://www.ncbi.nlm.nih.gov/nuccore/CP000645 CP000645]) which has an identical ''tniB''-GNAT junction as [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017'']; [https://tncentral.ncc.unesp.br/report/te/In34-AY123253 In34] ([https://www.ncbi.nlm.nih.gov/nuccore/AY123253 AY123253] ) <ref>{{#pmid:PMC149023}}</ref> is complex and, in addition, to carrying the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1326 IS''1326'']::[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1353 IS''1353''] insertion, also carries an insertion of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS26 IS''26'']-based transposon [https://tncentral.ncc.unesp.br/report/te/Tn4352-M20306 Tn''4352''] in ''tniA''; [https://tncentral.ncc.unesp.br/report/te/In_Tn1935-MK797990 In_Tn''1935''] ([https://www.ncbi.nlm.nih.gov/nuccore/MK797990 MK797990]) is identical to [https://tncentral.ncc.unesp.br/report/te/In34-AY123253 In34] except for its integron cassettes; and, finally, [https://tncentral.ncc.unesp.br/report/te/In_Tn5045-FN821089.1 In_Tn''5045''] ([https://www.ncbi.nlm.nih.gov/nuccore/FN821089 FN821089]) <ref name=":12" /> carries the ''tniB''-GNAT junction and also a [[Transposons families/Tn3 family|Tn''3''-family]] chromate resistance transposon, Tn [https://tncentral.ncc.unesp.br/report/te/TnOtChr.1-FN821089.1 Tn''OtChr.1''], at the 5’ end of ''tniB''. For [https://tncentral.ncc.unesp.br/report/te/In31-AJ223604 In31] ([https://www.ncbi.nlm.nih.gov/nuccore/AJ223604 AJ223604]) <ref>{{#pmid:PMC89222}}</ref>, ''tniB'' is truncated further to its 5’ end than those with a ''tniB''-GNAT junction and for [https://tncentral.ncc.unesp.br/report/te/In_Tn21.1-MH257753 In_Tn''21.1''] ([https://www.ncbi.nlm.nih.gov/nuccore/MH257753 MH257753]) an [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS26 IS''26''] insertion has led to a 3’ truncation of the ''tniA'' gene. | |||
A third group, which includes [https://tncentral.ncc.unesp.br/report/te/In33-AF313471 In33] ([https://www.ncbi.nlm.nih.gov/nuccore/AF313471 AF313471]) <ref name=":25">{{#pmid:PMC127177}}</ref>, [https://tncentral.ncc.unesp.br/report/te/In36-AY259085 In36] ([https://www.ncbi.nlm.nih.gov/nuccore/AY259085 AY259085]) <ref name=":15">{{#pmid:PMC161834}}</ref>, [https://tncentral.ncc.unesp.br/report/te/In4-U12338.3 In4] ([https://www.ncbi.nlm.nih.gov/nuccore/U12338.3 U12338.3]) <ref>{{#pmid:1648560}}</ref>, [https://tncentral.ncc.unesp.br/report/te/In37-AY259086 In37] ([https://www.ncbi.nlm.nih.gov/nuccore/AY259086 AY259086]) <ref name=":15" /> and [https://tncentral.ncc.unesp.br/report/te/In_Tn6025-GU562437 In_Tn''6025''] ([https://www.ncbi.nlm.nih.gov/nuccore/GU562437 GU562437]) have all undergone a complex rearrangement involving [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS6100 IS''6100''] and duplication of IRt and repeats in an inverted orientation surrounding the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS6100 IS''6100''] copy. | |||
The fourth group, including [https://tncentral.ncc.unesp.br/report/te/In7-L06418 In7] ([https://www.ncbi.nlm.nih.gov/nuccore/L06418 L06418])<ref name=":16">{{#pmid:PMC311882}}</ref>, [https://tncentral.ncc.unesp.br/report/te/In6_p-L06822 In6_p] ([https://www.ncbi.nlm.nih.gov/nuccore/L06822 L06822]) and [https://tncentral.ncc.unesp.br/report/te/InS21-AJ311891 InS21] are closely related but are only partially sequenced examples. They do not include an IRt end and include an insertion of IS''CR1'' ([[IS Families/IS91-ISCR families|IS91 and ISCR family]]). However, the reported partial integron [https://tncentral.ncc.unesp.br/report/te/In7-L06418 In7] ([https://www.ncbi.nlm.nih.gov/nuccore/KU997026 KU997026]) from ''[[wikipedia:Escherichia_coli|Escherichia coli]]'' plasmid pDGO100 <ref>{{#pmid:27318267}}</ref> closely resembles [https://tncentral.ncc.unesp.br/report/te/In34-AY123253 In34]. It extends both upstream to include the first [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS26 IS''26''] copy of transposon [https://tncentral.ncc.unesp.br/report/te/Tn4352-M20306 Tn''4352''] inserted in ''tniA'' of [https://tncentral.ncc.unesp.br/report/te/In34-AY123253 In34] and downstream to include the IRi end. The [https://tncentral.ncc.unesp.br/report/te/In7-L06418 In7] ([https://www.ncbi.nlm.nih.gov/nuccore/L06418 L06418]) copy <ref name=":16" /> is a sub-sequence of this. A similar problem is likely to arise with [https://tncentral.ncc.unesp.br/report/te/In6_p-L06822 In6_p] ([https://www.ncbi.nlm.nih.gov/nuccore/L06822 L06822]) <ref>{{#pmid:8378445}}</ref> which also shares several identical regions with [https://tncentral.ncc.unesp.br/report/te/In34-AY123253 In34]. [https://tncentral.ncc.unesp.br/report/te/In6_p-L06822 In6_p] includes a full set of IRi sequences downstream of its IntI1 gene but the sequence of its pSa plasmid parent <ref>{{#pmid:PMC217742}}</ref> is not available. Finally, [https://tncentral.ncc.unesp.br/report/te/InS21-AJ311891 InS21] ([https://www.ncbi.nlm.nih.gov/nuccore/AJ311891 AJ311891]), again with significant regions identical to [https://tncentral.ncc.unesp.br/report/te/In34-AY123253 In34], from plasmid [https://tncentral.ncc.unesp.br/report/te/InS21-AJ311891 pS21], carries a complete IRi end but is truncated at the opposite end. | |||
There is consequently no strong evidence that integron platforms along with their cassettes have been transferred without their accompanying [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] transposition system. “Integron” insertion may have occurred prior to decay of the ''tniABQR'' genes (see <ref name=":13" />). | |||
== Inter-transposon recombination at the Tn''402'' ''res'' == | |||
As in other transposons which use a site-specific resolution mechanism as a step in their transposition <ref name=":26">{{#pmid:PMC7157771}}</ref><ref name=":18">{{#pmid:22878084}}</ref>, members of the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 IS''402''] family can also undergo inter-transposon recombination at the ''res'' site and in this way exchange DNA modules. This type of behavior has been reported by Labbate et al. <ref name=":18" /><ref name=":19">{{#pmid:PMC2493286}}</ref> who observed that transposon [https://tncentral.ncc.unesp.br/report/te/Tn6007-EU591509.1 Tn''6007''] from ''[[wikipedia:Enterobacter_cloacae|Enterobacter cloacae]]'' included a class 1 integron with two non-antibiotic-resistance-type gene cassettes and a complete transposition module. The ''tniABQR'' module appeared to be a hybrid with a boundary within the ''res'' site ([[:File:Fig-Tn402.11.png|Fig. Tn402.11]]). The level of identity from the ''res'' site through ''tniR'' and the integron was observed to be high (99.9%) while that further upstream through the ''tniA'',''B'' and ''Q'' genes was only 89% <ref name=":19" />. This is not an isolated case and a number of additional examples of this phenomenon can also be observed ([[:File:Fig-Tn402.11.png|Fig. Tn402.11]]). Similar observations were made by Betteridge et al. in an analysis of integron platforms isolated from Human Commensal Bacteria in an '''I'''ntensive '''C'''are '''U'''nit (ICU) <ref name=":27">{{#pmid:PMC3147655}}</ref>. | |||
<br /> | |||
[[File:Fig-Tn402.11.png|center|thumb|680x680px|'''Fig. Tn402.11.''' Inter transposon recombination at res. [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] is shown as a pale yellow box. Open reading frames are shown as horizontal boxes where the arrowheads indicate the direction of translation. Lavender, transposase related genes; red, antibiotic resistance genes; purple, other; blue, IntI; the terminal IRs (IRt, transposase proximal; IRi, transposase distal) are also shown together with the short repeated sub-sites. The res site together with its six sub-repeats is shown in green as are the attI and attC recombination sites involved in acquiring integron cassettes. Above is shown the alignment of various [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] members (listed on the right). The extent of homology with different parts of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] are shown as red horizontal lines. The striped regions show regions of partial identity. These occur upstream orf and terminate precisely at, the res site Small triangles indicate insertions relative to [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']. The figure was generated using [https://www.snapgene.com/ SnapGene].]] | |||
== Origin of the Tn''402'' group integrons. == | |||
One hypothesis to explain the origin of the mobilizable class 1 integron is that it is derived from a chromosomal integron platform that became mobile by the acquisition of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']-like transposition functions. However, the identification of two other [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family groups with totally different passenger gene ensembles tends to suggest that it might have been an ancestral [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] transposon which acquired the passengers. | |||
The suggestion that some class 1 integrons identified in a sediment microbial community potentially predate integron association with [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] is difficult to assess <ref name=":13" />. Also, no ancestral [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] transposon has yet been identified without passenger genes. | |||
=== | === The Tn''Pfu1'' group === | ||
In the majority of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family transposons, '''''integron platform is of type 1''''' and the integrase gene is known as ''intI1''. ''IntI1'' is invariably located at the distal transposon end and is expressed in the same direction as ''tniA'', however, there is an additional group in which the integrase faces towards ''tniR''. | |||
An example of a [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] derivative with this configuration of ''tniA'' and ''intI'' genes, [https://tncentral.ncc.unesp.br/report/te/TnPfu1-LC331665.1 Tn''Pfu1''] ([https://www.ncbi.nlm.nih.gov/nuccore/LC331665.1 LC331665.1]), ([[:File:Fig-Tn402.11.png|Fig. Tn402.11]]) was first identified from ''Pseudomonas fulva'' and carried a ''bla''IMP-1 gene together with AAC(6') and ''[https://card.mcmaster.ca/ontology/36337 fosX]'' gene cassettes <ref name=":17" />. A BLAST search for ''tniA'' -''intI1'' regions with this configuration revealed only two additional examples, all closely related except for the gene cassettes carried by the integron platform: [https://tncentral.ncc.unesp.br/report/te/TnPfu1.1-LC589064 Tn''Pfu1.1''] ([https://www.ncbi.nlm.nih.gov/nuccore/LC589064 LC589064]) carries a ''bla'' GES gene and a cassette containing [https://card.mcmaster.ca/ontology/38979 AAC(6')-Ib8]/-Ib4, while [https://tncentral.ncc.unesp.br/report/te/TnPfu1.2-LC589062 Tn''Pfu1.2''] ([https://www.ncbi.nlm.nih.gov/nuccore/LC589062 LC589062]) includes two copies of ''bla'' GES. | |||
Members of this group carry another type of ''intI'' gene associated with [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']-like transposition genes, ''intI3''. This was first identified in ''[[wikipedia:Serratia_marcescens|Serratia marcescens]]'' plasmid pSMB731 as part of a new integron type, type 3 <ref>{{#pmid:PMC162793}}</ref>, in which the integrase showed only limited identity to IntI1. Unfortunately, only partial sequence data was presented ([[:File:Fig-Tn402.12.png|Fig. Tn402.12]]) although a limited extension was published more recently <ref>{{#pmid:PMC135066}}</ref> covering the ''tniA'' gene upstream and an [https://card.mcmaster.ca/ontology/38979 AAC(6')-Ib8 gene] cassette downstream together with the IRi end ([[:File:Fig-Tn402.12.png|Fig. Tn402.12]]). | |||
[[File:Fig-Tn402.12.png|center|thumb|680x680px|'''Fig. Tn402.12.''' '''The [https://tncentral.ncc.unesp.br/report/te/TnPfu1-LC331665.1 Tn''Pfu1''] members.''' [https://tncentral.ncc.unesp.br/report/te/TnPfu1-LC331665.1 Tn''Pfu1''] is shown as a pale yellow box. Open reading frames | |||
are shown as horizontal boxes where the arrowheads indicate the direction of translation. Lavender, transposase-related genes; red, antibiotic resistance genes; purple, other; blue, IntI ; the terminal IRs (IRt, transposase proximal; IRi, transposase distal) are also shown together with the short repeated sub-sites. The res site together with its six sub-repeats is shown in green, as are the attI and attC recombination sites involved in acquiring integron cassettes. Above is shown the alignment of the other Tn''Pfu1'' derivatives and of various DNA segments which identified the intI3 gene. The figure was generated using [https://www.snapgene.com/ SnapGene].]] | |||
<br /> | |||
=== The Tn''5053'' group === | |||
[https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] ([https://www.ncbi.nlm.nih.gov/nuccore/L40585.1/ L40585.1]) itself ([[:File:Fig-Tn402.4.png|Fig. Tn402.4]]), isolated from a Russian mercury mine <ref name=":0" />, carries an operon specifying resistance to mercury. The mercury resistance genes, like the antibiotic resistance genes in the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] group, are oriented such that expression occurs towards and downstream of the transposition genes. For the sake of simplicity, the '''IR''' distal to the transposition genes has retained its name from [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''], IRi. | |||
As for the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] and [https://tncentral.ncc.unesp.br/report/te/TnPfu1-LC331665.1 Tn''Pfu1''] groups, the TniA,B,Q and R proteins are well conserved ([[:File:Fig-Tn402.9a.png|Fig. Tn402.9]]) while the DNA sequences show a higher degree of variation ([[:File:Fig-Tn402.13.png|Fig. Tn402.13]]).[[File:Fig-Tn402.13.png|center|thumb|680x680px|'''Fig. Tn402.13.''' '''Alignment of Tn''5053'' group Tni regions against [https://tncentral.ncc.unesp.br/report/te/Tn5058-Y17897 Tn''5058''] showing sequence variation.''' Open reading frames are shown as horizontal boxes where the arrowheads indicate the direction of translation. Lavender, transposase-related genes; purple, other; the terminal IR (IRt, transposase proximal) is also shown together with the short repeated sub-sites. The res site with its repeated sequences is shown in green. Tn marked with '''*''' indicate those with a duplication of the mercury resistance operon. Above is shown the alignment of various [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053'']-related transposons (names listed on the right). The extent of homology with different parts of [https://tncentral.ncc.unesp.br/report/te/Tn5058-Y17897 Tn''5058''] are shown as red horizontal lines. Small triangles indicate insertions relative to [https://tncentral.ncc.unesp.br/report/te/Tn5058-Y17897 Tn''5058'']. The striped appearance indicates non-identical regions. | |||
The figure was generated using [https://www.snapgene.com/ SnapGene].]] | |||
=== Inter-transposon recombination at the Tn''5053'' ''res'' === | |||
Inter-transposon recombination at the ''res'' site is also observed in the case of family members which carry mercury resistance genes in place of the integron platform. This can be seen between [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] ([https://www.ncbi.nlm.nih.gov/nuccore/L40585.1/ L40585.1]) and a related mercury resistance transposon we have called [https://tncentral.ncc.unesp.br/report/te/Tn5053.1-CP002451 Tn''5053.1''] ([https://www.ncbi.nlm.nih.gov/nuccore/CP002451 CP002451]) from the ''[[wikipedia:Alicycliphilus|Alicycliphilus denitrificans]]'' BC plasmid pALIDE02 <ref>{{#pmid:PMC3692508}}</ref> ([[:File:Fig-Tn402.14.png|Fig. Tn402.14]]) where the tniABQ module has clearly been exchanged . This is would at least partially explain why [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] and [https://tncentral.ncc.unesp.br/report/te/Tn5053.1-CP002451 Tn''5053.1''] are located in different subgroups in the phylogenetic tree ([[:File:Fig-Tn402.8.png|Fig. Tn402.8]]) | |||
<br /> | |||
[[File:Fig-Tn402.14.png|center|thumb|680x680px|'''Fig. Tn402.14.''' Inter transposon recombination at res. [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] is shown as a pale yellow box. Open reading frames are shown as horizontal boxes where the arrowheads indicate the direction of translation. Lavender, transposase-related genes; yellow, mercury resistance genes; the terminal IRs (IRt, transposase proximal; IRi, transposase distal) are also shown together with the short repeated sub-sites. The res site together with its six sub-repeats is shown in green. Above is shown the alignment with [https://tncentral.ncc.unesp.br/report/te/Tn5053.1-CP002451 Tn''5053.1'']. The extent of homology is shown as red horizontal lines. The striped regions show regions of partial identity. Both transposons carry similar sets of mercury resistance genes (right) and a common tniR gene. Sequence identity breaks down immediately upstream of the res site. The figure was generated using [https://www.snapgene.com/ SnapGene].]] | |||
=== Presence of a Tn''21''-like IR === | |||
The relationship between members of this mercury resistance [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] group is complex. One close relative, [https://tncentral.ncc.unesp.br/report/te/Tn5053.4-CP001919 Tn''5053.4''] ([https://www.ncbi.nlm.nih.gov/nuccore/CP001919 CP001919]), carries a simple insertion of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS4321 IS''4321''] at the distal end. | |||
Interestingly, all mercury resistance [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family members carry a sequence resembling a [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21''] IR, suggesting that the mercury resistance genes may have be acquired from a [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21'']-derivative transposon towards the IRi end ([[:File:Fig-Tn402.15.png|Fig. Tn402.15]]) , suggesting that mercury resistance was originally acquired from a [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21'']-related [[Transposons families/Tn3 family|Tn''3'' family transposon]] (see <ref name=":28">{{#pmid:26350313}}</ref>). However, [https://tncentral.ncc.unesp.br/report/te/Tn5053.2-CP024682 Tn''5053.2''] ([https://www.ncbi.nlm.nih.gov/nuccore/CP024682 CP024682]) does not and is also missing the [https://www.uniprot.org/uniprot/P00392 MerA] sequence up to and including ''[https://www.uniprot.org/uniprot/P22874 merR]'' due to insertion of an ''ahp'' gene Moreover, in a subgroup, there has been a partial duplication of the mercury resistance operon followed by some decay and rearrangement with changes in gene number and order. This may have involved the [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21'']-like IR since these transposons, [https://tncentral.ncc.unesp.br/report/te/Tn5053.6-JX469833 Tn''5053.6''] ([https://www.ncbi.nlm.nih.gov/nuccore/JX469833 JX469833]), [https://tncentral.ncc.unesp.br/report/te/Tn5053.8-CP017991 Tn''5053.8''] ([https://www.ncbi.nlm.nih.gov/nuccore/CP017991 CP017991]), [https://tncentral.ncc.unesp.br/report/te/Tn5053.9-CP048650 Tn''5053.9''] ([https://www.ncbi.nlm.nih.gov/nuccore/CP048650 CP048650]), [https://tncentral.ncc.unesp.br/report/te/Tn5058-Y17897 Tn''5058''] ([https://www.ncbi.nlm.nih.gov/nuccore/Y17897 Y17897]) and [https://tncentral.ncc.unesp.br/report/te/Tn50580-AM048832 Tn''50580''] ([https://www.ncbi.nlm.nih.gov/nuccore/AM048832 AM048832]), all carry two [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21'']-like IR. | |||
[[File:Fig-Tn402.15.png|center|thumb|680x680px|'''Fig. Tn402.15.''' Alignment of [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] central region and right ends showing the [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21'']-like IR. Tn marked with * indicate those with a duplication of the mercury resistance operon. “Middle” indicates the internal IR-carrying region (boxed), “end” indicates the transposase distal end. Above is shown the alignment of various [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn5053]-related transposons (names listed on the right). The extent of homology with different parts of [https://tncentral.ncc.unesp.br/report/te/Tn5058-Y17897 Tn''5058''] are shown as red horizontal lines. The striped appearance indicates no- identical regions. | |||
The figure was generated using [https://www.snapgene.com/ SnapGene].]] | |||
The | <br /> | ||
=== Tn''5053'' group Mercury resistance genes === | |||
Mercury resistance genes are generally grouped into an operon. The major players are: ''merA'', encoding a mercuric reductase (MerA), which catalyses reduction of Hg(II) to volatile Hg(0) and is the central enzyme in Hg(II) resistance <ref>{{#pmid:20545753}}</ref><ref>{{#pmid:PMC3466566}}</ref>; ''merB'' encoding an organomercury lyase (MerB); ''merP'' encoding a periplasmic Hg(II) scavenging protein (MerP); ''merT'', ''merC'', ''merE'', ''merF'' and ''merG'' which encode inner membrane spanning proteins (MerT, MerC, MerE, MerF, MerG) that transport Hg(II) to the cytoplasm where it is reduced by MerA; ''merR'' and ''merD'' encoding the regulatory proteins (MerR, MerD). MerR is a transcriptional repressor in the absence of Hg(II) and an activator in its presence and MerD downregulates the operon (for review see <ref>HAMLET N, LANDALE E, DAVIS B, SUMMERS A. Roles of the Tn2l merT, merP, and merC Gene Products in Mercury Resistance and Mercury Binding. J Bacteriol. 1992;174:6377–6385.</ref>) . | |||
Mercury resistance operons do not all carry a complete set of these genes. [[:File:Fig-Tn402.16.png|Fig. Tn402.16]] shows a sample of mercury resistance operons from various transposons of the [[Transposons families/Tn3 family|Tn''3'']] and Tn''402'' families together with the ensemble of ''mer'' genes in the order in which they occur in the canonical [[Transposons families/Tn3 family|Tn''3'' family transposon]] [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21'']. | |||
[[File:Fig-Tn402.16.png|center|thumb|800x800px|'''Fig. Tn402.16.''' '''Mercury Resistance Genes in the [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] Group.''' The table lists each of the different genes involved in mercury resistance using the order observed in the well-studied Tn''21'' transposon. Mercury resistance transposons are listed in the left-hand column. Those marked '''*''' are members of the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family. Resistance gene order is shown in the top line. | |||
Those corresponding to the [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21''] order are marked in black. Those which have been rearranged are shown in red. _p indicates that the transposon is incomplete (partial).]] | |||
== Insertion Sites: ''res'' Hunters == | |||
In early studies describing [https://tncentral.ncc.unesp.br/report/te/Tn502-EU306743 Tn''502''] and its differences with other mercury resistance transposons known at the time <ref name=":29">Stanisich V, Arwas R, Bennett PM, De La Cruz F. Characterization of Pseudomonas Mercury - resistance Transposon Tn502, Which Has a Preferred Insertion Site in RP1. J Gen Microbiol. 1989;135:2909–2915.</ref> it was observed that it displays a preference for insertion into a single ''PstI'' fragment of the plasmid RP1 <ref name=":23" />. Further restriction mapping led to the conclusion that the insertions occurred at the same site, largely in one orientation. Deletion of this region resulted in a marked reduction in [https://tncentral.ncc.unesp.br/report/te/Tn502-EU306743 Tn''502''] insertion frequency into the plasmid <ref name=":29" />. This region was subsequently shown to carry the non-transcribed spacer of the ''par'' locus of plasmid RP1, initially defined in the related plasmids RK2 and RP4 as a region required for plasmid stabilisation <ref>{{#pmid:PMC526801}}</ref><ref>{{#pmid:PMC526800}}</ref>. It is similarly organised to the ''res'' region of [[Transposons families/Tn3 family|Tn''3'' family transposon]] (see <ref name=":26" /><ref name=":28" />) and contains three binding sites for the ParA site-specific recombinase (''resI'', ''resII'' and ''resIII'' ) <ref>{{#pmid:8057833}}</ref>. | |||
Tn''2521'', a multi-resistant transposon identified in the chromosome of Pseudomonas aeruginosa clinical isolates in 1982 | A similar sequence-specific insertion of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']/[https://tncentral.ncc.unesp.br/report/te/Tn5090-AM993098 Tn''5090''] into derivatives of plasmids R388 and RP1 was also demonstrated experimentally <ref name=":30">{{#pmid:10779712}}</ref>. In RP1, insertions occurred in two major clusters ([[:File:Fig-Tn402.17a.png|Fig. Tn402.17]] '''A''') within 140 bp: one cluster (1) was located in the ''resII'' resolvase subsite and the second (2) within the ''parC'' gene. The orientation of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] insertions in each cluster was inverted. A single major insertion site was observed in plasmid R388, again, close to the proposed ''res'' sites <ref name=":30" />. Furthermore, this targeting required an intact ParA since partial deletion significantly reduced the attractiveness of the derivative plasmid as a target. Moreover, a plasmid in which the ParA active site serine was mutated completely retained its activity as a transposition target <ref name=":30" /> showing that it is the binding of resolvase and presumably the architecture of the ''res''-ParA nucleoprotein complex rather than recombination activity which determine target activity.<br />[[File:Fig-Tn402.17a.png|center|thumb|800x800px|'''Fig. Tn402.17.''' '''Insertion sequence specificity. Res subsequences are shown in green boxes.''' Vertical arrowheads show individual insertions. These are colored according to the different inserted TE. '''A)''' insertions of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] into and near the ''par'' site of plasmid RP1 which functions in assuring separation of plasmid dimers and their correct segregation <ref name=":9" />. '''B)''' Insertions of different family members into the par site of RP1 ([https://www.ncbi.nlm.nih.gov/nuccore/BN000925 BN000925]). [https://tncentral.ncc.unesp.br/report/te/Tn502-EU306743 Tn''502'']; Tn''502''DtniQ; [https://tncentral.ncc.unesp.br/report/te/Tn512-EU306744 Tn''512'']; [https://tncentral.ncc.unesp.br/report/te/In0-U49101 In0]; [https://tncentral.ncc.unesp.br/report/te/In2-AF071413 In2] <ref name=":34" />.]] | ||
[https://tncentral.ncc.unesp.br/report/te/In33-AF313471 Tn''2521''], a multi-resistant transposon identified in the chromosome of ''[[wikipedia:Pseudomonas_aeruginosa|Pseudomonas aeruginosa]]'' clinical isolates in 1982 <ref name=":31">{{#pmid:PMC220297}}</ref> was also observed to have marked integration site-specificity in transposition into IncP plasmids (in this case R18-1, identical to R68). [https://tncentral.ncc.unesp.br/report/te/In33-AF313471 Tn''2521''] was later shown to be an integron (In33) without ''tni'' genes and its insertion point in the R18-18::[https://tncentral.ncc.unesp.br/report/te/In33-AF313471 Tn''2521''] example examined was shown to have inserted at the R18-18 equivalent to RP1/RP4 site ''resII'' <ref name=":25" />. Moreover, additional analysis of the flanking sequences of other integron structures indicated that insertion occurred in the ''res'' sites of several [[Transposons families/Tn3 family|Tn''3'' family transposon]] <ref>{{#pmid:PMC90776}}</ref>: [https://tncentral.ncc.unesp.br/report/te/In28-AF313472 In28] in the ''resI'' region of Tn''1403''; [https://tncentral.ncc.unesp.br/report/te/In4-U12338.3 In4] abutting ''resI'' in [https://tncentral.ncc.unesp.br/report/te/Tn1696-U12338.3 Tn''1696'']; In1 upstream of ''resP'' in R46 ( <ref>{{#pmid:6264522}}</ref>, also called TP120 <ref>{{#pmid:325391}}</ref>), the region expected to include the plasmid ''res'' site which has an adjacent functional resolvase-like gene, ''uvp1'' <ref>{{#pmid:9648746}}</ref>, and is similar to and can undergo recombination with that of [https://tncentral.ncc.unesp.br/report/te/Tn1-NC_008357 Tn''1''] <ref>{{#pmid:6306704}}</ref><ref name=":35">{{#pmid:3020154}}</ref><ref>Dodd HM, Bennett PM. The R46 Site-specific Recombination System is a Homoiogue of the Tn3 and gamma delta (Tn1000) Cointegrate Resolution System.</ref>. In these examples, the inserted integron platform does not contain the set of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] transposition enzymes. This implies either that transposition preceded decay of the transposition genes or that their products were provided ''in trans'' from another transposon in the cell as is suggested by transposition of [https://tncentral.ncc.unesp.br/report/te/In33-AF313471 In33] ([https://tncentral.ncc.unesp.br/report/te/In33-AF313471 Tn''2521'']). A further example, however, is the insertion into [https://tncentral.ncc.unesp.br/report/te/Tn1721-X61367.1 Tn''1721''] of an integron with a full complement of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] transposition enzymes ([https://www.ncbi.nlm.nih.gov/nuccore/HQ730118 HQ730118]) <ref name=":27" />. | |||
This type of insertion site-specificity was also observed for Tn''5053'' | This type of insertion site-specificity was also observed for [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] <ref name=":0" />, again using RP1 and an RP4 derivative plasmid target. Sequence analysis showed that the insertions were flanked by a 5 bp direct target repeat as expected <ref name=":0" /><ref name=":22" />. A survey of conjugative plasmids (cited in <ref name=":32">{{#pmid:10476039}}</ref>) identified only a single example which acted as an efficient [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] target. This plasmid carried Tn''701'', a close relative of Tn''1721'' and The [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] insertion were reported to have occurred in the ''res'' site of this transposon. Introduction of Tn''701'' or [https://tncentral.ncc.unesp.br/report/te/Tn1721-X61367.1 Tn''1721''] into plasmids not generally active targets such as R387, pOXGen or pBR322, these became efficient [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] targets <ref name=":32" />. In addition, a database search using the first 50 bp of the ends of [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] and [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] as the query sequences revealed a number of Tn junctions in which the IRi ends were located upstream of resolvase-like genes <ref name=":32" />. | ||
In these studies | In these studies <ref name=":32" />, Insertion of Tn''5035'' was observed experimentally using as target the cloned insertion-free ''res'' site of [https://tncentral.ncc.unesp.br/report/te/Tn5044-Y17691.1 Tn''5044''] <ref>{{#pmid:10875286}}</ref> together with the native TnpR gene. Very strong clustering was observed between ''resI'' and ''resII'' and all but one insertion occurred in the same orientation ([[:File:Fig-Tn402.17b.png|Fig. Tn402.17]] '''C'''). Moreover, when cloned and in the absence of TnpR, the ''res'' sites of [https://tncentral.ncc.unesp.br/report/te/Tn1721-X61367.1 Tn''1721''] <ref>{{#pmid:1312499}}</ref>, [https://tncentral.ncc.unesp.br/report/te/Tn5036-Y09025 Tn''5036''] <ref name=":21" />, Tn''5051'' (no sequence available) <ref name=":21" /><ref name=":32" /> and Tn''5059'' ([https://www.ncbi.nlm.nih.gov/nuccore/Y10102 Y10102, partial sequence]) were not attractive to Tn''5035'' insertion, however, when TnpR from [https://tncentral.ncc.unesp.br/report/te/Tn1721-X61367.1 Tn''1721''] was supplied in trans, efficient targeted insertion occurred and sequence analysis showed strong clustering ([[:File:Fig-Tn402.17a.png|Fig. Tn402.17]] '''D'''). This behavior rules out the possibility that expression (transcription or translation) of TnpR in its natural proximity to res provides a suitable integration substrate. A similar requirement for the ParA resolvase was reported for insertion into the RP1 res. Surprisingly target attractiveness appeared to be limited to the [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21''] branch of the [[Transposons families/Tn3 family|Tn''3'' family]] <ref name=":20" /><ref>{{#pmid:6312271}}</ref> neither [https://tncentral.ncc.unesp.br/report/te/Tn1-NC_008357 Tn''1''] nor [https://tncentral.ncc.unesp.br/report/te/Tn1000-X60200.1 Tn''1000''] were found to be efficient targets for [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] insertion. | ||
Tn''402'' family members with mercury resistance genes, Tn''502'' and Tn''512'', have also been found to show quasi-identical insertion specificity in an RP1 res target | [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family members with mercury resistance genes, [https://tncentral.ncc.unesp.br/report/te/Tn502-EU306743 Tn''502''] and [https://tncentral.ncc.unesp.br/report/te/Tn512-EU306744 Tn''512''], have also been found to show quasi-identical insertion specificity in an RP1 res target <ref name=":24" /> as [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] itself ([[:File:Fig-Tn402.17a.png|Fig. Tn402.17]] '''B'''). Remarkably, the insertion site observed in resII was identical to that observed for [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']/[https://tncentral.ncc.unesp.br/report/te/Tn5090-AM993098 Tn''5090''] (cluster 1 in [[:File:Fig-Tn402.17a.png|Fig. Tn402.17]] '''A''')<ref name=":30" />. The authors also address the potential mobility of integron platforms, such as [https://tncentral.ncc.unesp.br/report/te/In0-U49101 In0] ([https://www.ncbi.nlm.nih.gov/nuccore/U49101 U49101]) and [https://tncentral.ncc.unesp.br/report/te/In2-AF071413 In2] ([https://www.ncbi.nlm.nih.gov/nuccore/AF071413 AF071413]) which do not contain a full set of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] ''tni'' genes but carry both IRi and IRt ends and are found with flanking 5 bp direct target repeats. Transposition of [https://tncentral.ncc.unesp.br/report/te/In2-AF071413 In2] from its location in [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21''] and of [https://tncentral.ncc.unesp.br/report/te/In0-U49101 In0] from its location in plasmid pVS1 was tested by supplying [https://tncentral.ncc.unesp.br/report/te/Tn502-EU306743 Tn''502''] ''tni'' genes ''in trans''. Transposition occurred at significant frequencies if both ''tni'' and the RP1 ''par'' site were present. Moreover, the vast majority of insertions occurred in the expected orientation at the cluster 1 site ([[:File:Fig-Tn402.17a.png|Fig. Tn402.17]] '''B'''). One puzzling observation was that although neither [https://tncentral.ncc.unesp.br/report/te/In0-U49101 In0] nor [https://tncentral.ncc.unesp.br/report/te/In2-AF071413 In2] include a [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] res site never-the-less, the transposition products had undergone resolution. | ||
Complementation by a resident Tn''402'' family transposon would provide an explanation for transposition of Tn''2521'' (In33) into the R18-1 | Complementation by a resident [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family transposon would provide an explanation for transposition of [https://tncentral.ncc.unesp.br/report/te/In33-AF313471 Tn''2521''] ([https://tncentral.ncc.unesp.br/report/te/In33-AF313471 In33]) into the R18-1 <ref name=":31" /> ''res'' site even though it does not contain a single [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] transposition gene <ref name=":25" />. | ||
In summary, all members of the Tn''402'' family tested appear to target ''res'' sites of plasmids and Tn''3'' family transposons of the Tn''21'' group. They require the cognate resolvase protein | In summary, all members of the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family tested appear to target ''res'' sites of plasmids and [[Transposons families/Tn3 family|Tn''3'' family transposons]] of the [https://tncentral.ncc.unesp.br/report/te/Tn21-AF071413 Tn''21''] group. They require the cognate resolvase protein <ref name=":30" /><ref name=":32" /> but do not require it to be catalytically active <ref name=":30" />. The role of TnpR could be structural, providing an architecturally precise nucleoprotein complex for integration and/or furnishing an interface for direct protein-protein interaction with a TniABQ component. Transposons with ''tni'' gene sets can also complement ''in trans'' integron platforms which are lacking these genes <ref name=":24" /> in principal providing the capacity to mobilise non-autonomous integron structures. | ||
<br /> | <br /> | ||
[[File:Fig-Tn402.17b.png|center|thumb|800x800px|'''Fig. Tn402.17.''' '''Insertion sequence specificity. Res subsequences are shown in green boxes.''' Vertical arrowheads show individual insertions. These are colored according to the different inserted TE. '''C)''' Insertions of [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] into the res site of [https://tncentral.ncc.unesp.br/report/te/Tn5044-Y17691.1 Tn''5044''] <ref name=":35" />. '''D)''' Insertions of [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] into the [[Transposons families/Tn3 family|Tn''3'' family res site]], and insertions into res sites of [https://tncentral.ncc.unesp.br/report/te/Tn1721-X61367.1 Tn''1721''], Tn''5051'', and Tn''5059'' with TnpR ([https://tncentral.ncc.unesp.br/report/te/Tn1721-X61367.1 Tn''1721'']) supplied in trans; [https://tncentral.ncc.unesp.br/report/te/Tn5053-L40585.1 Tn''5053''] insertions into plasmids with intact res site and tnpR gene of [https://tncentral.ncc.unesp.br/report/te/Tn1721-X61367.1 Tn''1721''] or [https://tncentral.ncc.unesp.br/report/te/Tn5036-Y09025 Tn''5036''].]] | |||
== A reflection on transposition mechanism. == | |||
Except for the bioinformatic analysis of their transposition proteins and the demonstration that their transposition requires a cointegration step which is resolved at a specific site, the ''res'' site and involves the TniR site-specific recombinase | Except for the bioinformatic analysis of their transposition proteins and the demonstration that their transposition requires a cointegration step which is resolved at a specific site, the ''res'' site and involves the TniR site-specific recombinase <ref name=":8" />, there has been no significant biochemical or structural studies of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family transposition. | ||
Studies with purified TniA | Studies with purified TniA <ref name=":9" /> have shown that the protein binds to the left IRt end of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']/[https://tncentral.ncc.unesp.br/report/te/Tn5090-AM993098 Tn''5090''] to generate six retarded bands (I-VI). This was further explored using DNase footprinting which showed that the four sites t1-t4 were Species I-III or of the resolvase TniR to the repeated sequences at the ''res'' site. It is therefore not known which of the multiple end sites are important for transposition or whether all repeated sequences in and around the proposed crossover point between r1 and r2 are necessary for resolution. | ||
or of the | Neither have the roles played by TniA, TniB or TniQ been determined. The fact that TniA carries a DDE motif and shows 25% similarity to TnsB transposase <ref name=":33">{{#pmid:26104363}}</ref> of [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 transposon Tn''7''] <ref name=":6" /><ref name=":8" /> strongly suggests that it is the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family transposase and the observation that TniB shares similarities with TnsC suggests that it may play a DNA-dependent bridging reaction between TniA and TniQ. It seems possible that, like TnsD and TnsE of [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] <ref name=":33" /><ref>{{#pmid:PMC312648}}</ref><ref>{{#pmid:PMC145423}}</ref>, ''tniQ'' is involved in the choice of the target sequence. Binding to the small repeated sequences at IRt and IRi also has yet to be demonstrated. | ||
In view of the sequence similarities between the [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] and [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family transposition proteins, it is interesting to note that the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] family possesses a dedicated system to resolve cointegrates whereas [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] does not. In contrast, [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] encodes two endonucleases, TnsA and TnsB. The DDE transposase, TnsB, binds to the [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] ends cleaves only one strand of the transposon whereas TnsA, which resembles a restriction endonuclease <ref>{{#pmid:10911996}}</ref>, cleaves the complementary strand. Together, both, form a heteromeric complex and in concert, achieve cleavage and joining reactions at both Tn''7'' ends <ref>{{#pmid:PMC452457}}</ref>. Transposition of Tn''7'' normally occurs by a cut-and-paste mechanism <ref>{{#pmid:1645619}}</ref>. However, although TnsA is required for TnsB activity, mutation of the active site of TnsA, prevents the '''cut-and-paste pathway''' and results in the formation of cointegrates <ref>{{#pmid:8602527}}</ref>. Therefore, it seems probable that TniA, like TnsB, catalyses only single-strand cleavage at each transposon end and to deal with the resulting cointegrate structures, [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] has recruited a resolvase-like protein and an accompanying abutting res site while [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] has recruited a second restriction endonuclease protein. | |||
An additional observation is that the majority of [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']-family transposons and integrons are flanked by 5 bp direct target repeats. This implies that intermolecular transposition represents a significant, if not unique pathway since intramolecular transposition using a cointegrate pathway leads to loss of the direct target repeats [[:File:1.23.1.png|(Fig.17.1]] and [[:File:1.23.2.png|Fig.17.2)]] | |||
== Acknowledgements == | |||
We would like to thank [https://www.latrobe.edu.au/physiology-anatomy-and-microbiology/research/petrovski Steve Petrovski] ([https://www.latrobe.edu.au/ La Trobe University], Australia) for critical comments. | We would like to thank [https://www.latrobe.edu.au/physiology-anatomy-and-microbiology/research/petrovski Steve Petrovski] ([https://www.latrobe.edu.au/ La Trobe University], Australia) for critical comments. | ||
==Bibliography== | ==Bibliography== | ||
< | {{Reflist|32em}} | ||
== How to Cite? == | |||
TnPedia Team. (2025). TnPedia – Chapter: The Tn''402'' Family of Prokaryotic Transposons. Zenodo. https://doi.org/10.5281/zenodo.15566551 | |||
[[File:Tn402-zenodo.15566551.png|link=https://doi.org/10.5281/zenodo.15566551|DOI badge]]<hr> | |||
{{TnPedia}} | |||
Latest revision as of 20:01, 31 May 2025
Historical
Tn402 was first identified [1] as an insertion of trimethoprim resistance from the IncP plasmid R751 [2][3] into a bacteriophage lambda derivative phage. Historically, this family of Tn have been intimately linked to integrons and, indeed, Tn402-like transposons have proved instrumental in integron dispersal. This first became apparent during the study of integron structures which include an integron integrase gene, intI, and various antibiotic resistance gene cassettes. These structures were found in different sequence environments suggesting that they are mobile genetic elements. Since they did not contain typical transposition-related genes or obvious terminal inverted repeats, it was suggested that integrons were unusual mobile elements distinct from typical transposons [4]. However, identification of an integron structure which is embedded in the Tn3 family transposon, Tn21, and flanked by 25bp Inverted Repeats (IR), in turn flanked by 5bp Direct Repeats (DR), suggested that the integron was part of a precursor transposon [5][6] whose transposition genes may have decayed following insertion into Tn21.
Indeed, integrons In0, In2, and In5 were later identified as defective transposon derivatives [7]. This clearly raises the question of how to define and name integrons (see Integrons). We will use a working definition for an integron as a DNA segment composed of an “integron platform” together with a variable number of integron gene cassettes. The integron platform includes an integrase gene, intI1 (a tyrosine site-specific recombinase), and a recombination site just upstream (attI) at which integron cassettes can be integrated. The cassettes are integrated in an orientation which allows their expression from a promoter, Pc, located within the integrase gene itself (Fig. Tn402.1) [8][9]. This definition is restrictive since it does not include the Tn402 ends of the transposition enzymes. It raises the important question of how transposable elements such as Tn402 and a second transposon family, Tn7, members have acquired these gene cassette-acquiring systems.
Tn402 (U67194.4) is similar if not identical to Tn5090 (AM993098) also isolated from R751[10]. Both had subsequently been grouped into a family called Tn5053 [11] although we have elected here to call the family the Tn402 family based on its founding transposon.
More recently, a second group of related transposons, the TnPfu1 group, was identified which include another integrase gene, IntI3, oriented in the opposite direction to that in the Tn402 group [12], and therefore acquired by a Tn402 family independently of the type 1 integron, while a third group, Tn5053, include genes for resistance to mercury salts instead of the integron platform [13].
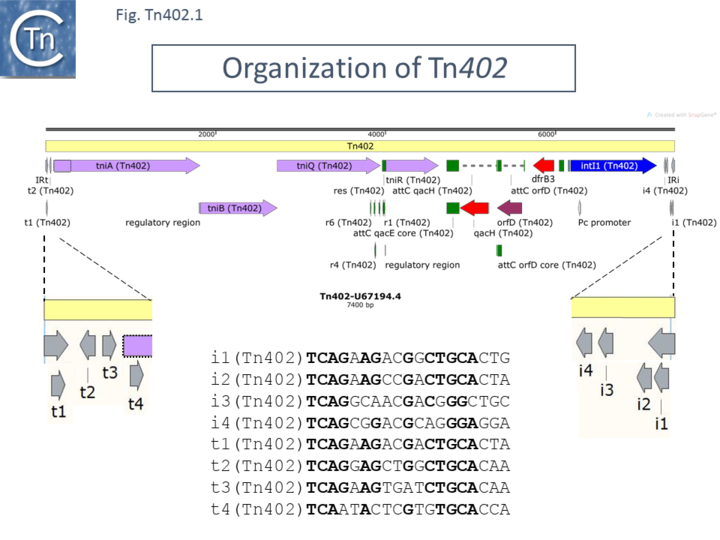
Distribution
Tn402 family transposons are widely distributed in clinical and environmental isolates. A number of community studies have been carried out including from aquatic environments [14], permafrost [15][16][17][18], the rhizosphere [19][20] and river and sewage sediments [21][22]. Because of their importance in environmental pollution, there has been much attention given to transposons carrying mercury resistance genes (see [23]). These have been identified, for example, as part of plasmids isolated from the river Mersey (U.K. [14]) in 1988 where an example was shown to share the same strict target specificity (Insertion Sites: res Hunters) and carry similar mercury resistance genes [24] to Tn5053, isolated from a Russian mercury mine [13] in 1984.
A derivative, Tn50580 carrying a complex set of mercury resistance genes was also identified in samples taken from sediment of the Nura, a highly contaminated river in Kazakhstan [22]. Other examples include, related transposons and more complex Tn5053 derivatives in permafrost samples many thousands of years old [15]; variants or transposon fragments with identical restriction maps (or partial sequences) in pre-antibiotic era strains in the Murray collection [25] (Enterobacteriaceae isolated between 1917 and 1954 [26]); association with plasmids also involved with degradation of the herbicide atrazine [27]; and in a variety of bacteria including Pseudomonas putida, Escherichia coli, Enterobacter and Klebsiella from water and from soil from various geographical regions mainly, but not exclusively in Russia [28].
Interestingly, Tn402-like transposons and transposon fragments were identified in a survey of IncP group plasmids identified in the rhizosphere of greenhouse-grown plants [19]. Two included drug resistance gene cassettes but none were associated with mercury resistance. On the other hand, a study of an alfalfa rhizosphere in Germany revealed a number of plasmids carrying mercury resistance among which was pSN102 which carried the Tn402-family transposon Tn5718 [20][22].
In view of the overuse of antibiotics, it is perhaps not surprising that Tn402 derivatives carrying antibiotic resistance integron cassettes have been found in sewage treatment plants (e.g. TnpRSB105 carried by plasmid pRsB105;[29]) and also as rearranged fragments (e.g.[30][31][32]) associated with other (rearranged TE) and also in estuarine waters [33].
Organization
Tn402 Terminal Repeated Sequences
Since it is now known that Tn402 and Tn5090 are different isolates of the same element and given that Tn402 was the first described, we have elected to retain the name Tn402 throughout although it should be kept in mind that in some of the literature, the transposon is described as Tn5090. The nucleotide sequence of Tn402 revealed a 7.4 kb region from R751 flanked by two 25bp IRs (Fig. Tn402.1). Two segments of Tn402 including each of the ends were found to be similar to the integron identified in the Tn3 family transposon, Tn21 [34], and called Tn5092, and to a DNA segment embedded in Tn5086 called Tn5093 [10][35].
In all cases, the IR were flanked by 5 bp direct target repeats different for each Tn. All three Tn included a type I integron integrase intI1 at one end. The IRs are referred to as IRi for the copy proximal to the integrase end and IRt for the copy proximal to the opposite (transposase) end. The transposon ends also include several 19 bp repeats: t1, t2 and t3 located at the IRt end with t2 oriented inversely and i1, i2, i3 and i4 located at the IRi transposon end (Fig. Tn402.1). A fourth repeat, t4, can be identified by inspection. These are binding sites for the transposase [36] and their differential organisation at each end would serve to distinguish between the ends in the transposition process as has been shown for transposon Tn7 [37][38][39] and bacteriophage Mu [40][41] (Fig. Tn402.2) and members of the IS21 family [42].
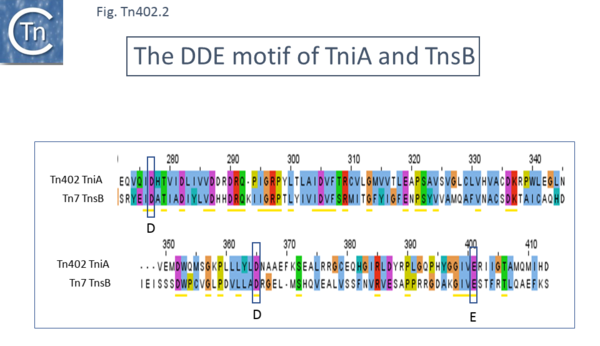
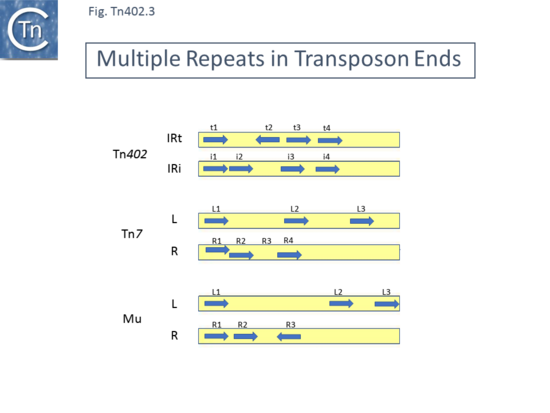
Tn402 Integron Cassettes
Tn402 carries three integron cassettes. One of these contains the trimethoprim resistance gene, dhfrIIc (dihydrofolate reductase type IIc) originally identified in the first transposition experiments [1]. This is located proximal to the integron platform promoter (Fig. Tn402.1). Another, orfD, of unkown function but observed in other integrons, is located downstream and this is followed by a gene encoding the exporter, qacE/qaceH.
Tn402 Transposition-Related Genes
Four transposition-related orfs were also identified. Three of these, tniA, tniB and orf6, now called tniQ, [11] specify proteins of 559, 302 and 405 amino acids respectively. They occur in that order and are all oriented in the same direction (Fig. Tn402.1). They probably constitute an operon expressed from a predicted single promoter [10]. The fourth gene, initially called tniC, but now renamed as tniR (see below) encodes a 207 amino acid protein.
Tn402 Transposition-Related Genes: tniA
TniA was predicted to be a transposase since it shows some weak similarity to the transposon Tn7 transposase TnsB (Fig. Tn402.2), and to the Tn552 transposase (25% identity) and includes a DDE motif typical of the catalytic site of many transposases. tniA is located close to IRt. Transposon Tn7, TnsB, which binds to the multiple short 22bp repeated sequences located in the left and right Tn7 ends (Fig. Tn402.3), suggesting that TniA may bind to the 19 bp repeated sequences in the Tn402 ends. This has now been demonstrated using DNase and hydroxy-radical footprinting approaches [36]. It should be noted that there is some ambiguity in the position of the TniA initiation codon (see below; Fig. Tn402.6).
Tn402 Transposition-Related Genes: tniB
The product of the second orf, TniB (Fig. Tn402.1), contains predicted nucleotide triphosphate binding sites and was proposed to be involved in nucleotide triphosphate hydrolysis [10]. It exhibits 21% identity to the Tn7 TnsC protein, an ATPase [45] which binds non-specifically to DNA in the presence of ATP [46], and is required for transposition. TnsC also binds to the TnsB component the heterodimeric TnsA/TnsB transposase and couples it to a protein involved in targeting Tn7 insertion, TnsD [47]. TniB also shows 24% identity to the MuB protein, also a DNA-dependent ATPase that preferentially stimulates intermolecular DNA strand transfer in bacteriophage Mu [48].
Tn402 Transposition-Related Genes: tniQ
The third gene (Fig. Tn402.1) was called orf6 [10] but has since been named tniQ [11] and its product did not exhibit similarity to known proteins [10]. It is positioned such that its initiation codon overlaps the termination codon of the upstream tniB gene (see below; Fig. Tn402.7) indicating that expression of the two gene products is translationally coupled.
Tn402 Transposition-Related Genes: tniR
The product of the fourth gene belongs to the invertase/resolvase family of site-specific recombinases. It has significant identity (67%) to the resolvase of the Tn3 family transposon, Tn5393, 47% identity to the Gin recombinase of bacteriophage Mu [49]. For this reason, it is now generally referred to as tniR for Resolvase. It is probably expressed separately from the other three genes and appears to have its own promoter [10] (see below; Fig. Tn402.5 A).
Tn5053 Terminal Repeated Sequences
More information was obtained from the related Tn5053 (L40585.1), a mercury resistance transposon isolated from the chromosome of a mercury resistant Xanthomonad from a mercury mine [13]. Tn5053 carries a set of transposition genes which are very similar to those of Tn402/Tn5090. Like other members of the family it is bordered by 25 bp inverted repeats (Fig. Tn402.4) and includes short ~19 bp repeat sequences at its ends.
Careful inspection of the Tn5053 sequence identifies additional 19bp repeat sequences than those reported [13], as in the case of Tn402/Tn5090 (Fig. Tn402.1). In this case there are 4 copies at the IRt end and three at the equivalent of the IRi end. Also, in the original article, it appears that those at the IRt and IRi ends have been exchanged compared to Tn402/Tn5090. Tn5053 carries a full complement of transposition genes tniA,B,Q and R but, instead of the integron recombination platform and antibiotic resistance gene cassettes, the transposon carries a mercury resistance operon (Fig. Tn402.4).
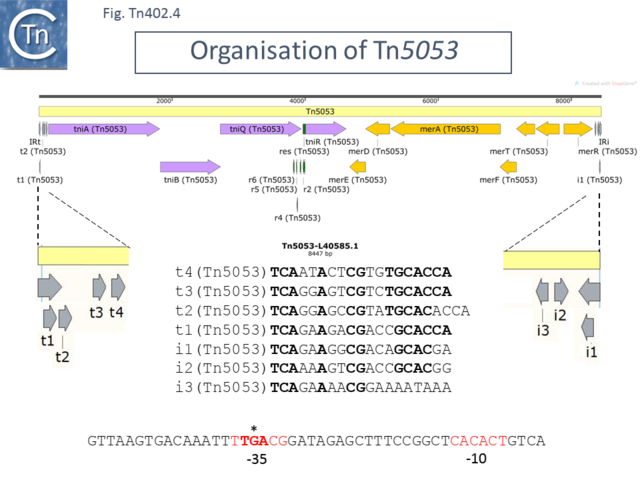
Tn5053 Functional analysis of Transposition-Related Genes
Transposition of a collection of insertion and deletion mutants of Tn5053 introduced into tniA, tniB and tniQ was tested in a conjugation (mating out [50]) assay using an RP4 plasmid derivative as a target [11]. Transposition was undetectable in all mutants but could be complemented by supplying a set of Tn5053 genes in trans from a third compatible plasmid.
It was also shown that the set of three genes from Tn402 were capable of complementing the mutant Tn5053 derivatives [11]. To address whether the mutations may have a polar effect on the downstream genes, mutations in the complementing set of genes were used: a Tn5053 tniA mutant could be complemented by a tniA operon with a wildtype tniA and carrying mutations in tniB and tniQ , while a tniB mutant could be complemented by a tniQ/tniR mutant. It is unclear whether a tniB mutant could be complemented by a tniA/tniQ/tniR mutant raising the formal possibility that some polarity effects may occur.

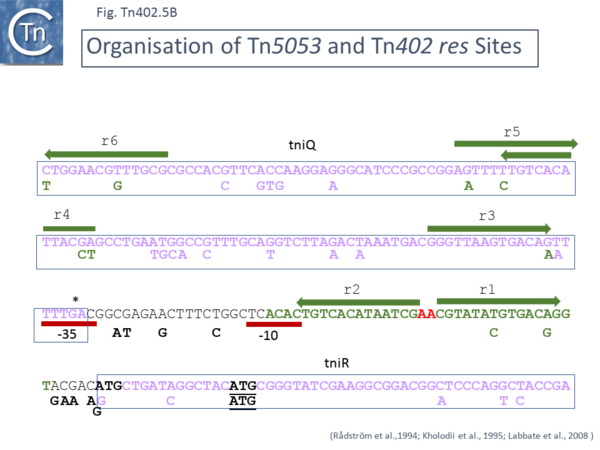
Tn5053 Functional Analysis of TniR and the res site
As might be expected from the similarity of tniR to site-specific resolvases, Tn5053 appears to transpose via a cointegrate intermediate. Ablation of the tniR gene and part of the tniQ gene resulted in a transposon unable to undergo transposition, presumably because of the defective TniQ. Complementation using the entire set of transposition genes resulted in high transposition levels. In the absence of TniR in the complementing plasmid, cointegrates carrying both donor and recipient plasmid antibiotic resistance markers were identified. Resolution required the presence of only TniR and not TniA, B or Q.
The recombination site was located using a system with cloned Tn restriction fragments from a region upstream of tniR and the “kinetics“ of resolution was followed. It was concluded that a region close to the 5’ end of the tniR gene which, on its own resulted in slow resolution, represented the principal resolution site while a region stretching upstream into the 3’ end of the tniQ gene was required for robust activity and therefore presumably carried secondary stimulating sequences. This region includes six 14 bp repeat sequences with the consensus PyTGTCACPuT(NT)(NT)NC(C/G), labelled r1-r6 (shown in detail in Fig. Tn402.5 A and Fig. Tn402.5 B).
Most site-specific resolution systems include multiple short repeated sequences which serve as recombinase binding sites permitting the assembly of architecturally precise nucleoprotein complexes to tightly control recombination. The repeats include r1 and r2 which are inverted, almost abut the start of the tniR gene and are separated by a dinucleotide, AA, proposed as the crossover point in the resolution recombination reaction (Fig. Tn402.4). Additional copies are observed further upstream and located within the 3’ end of tniQ (Fig. Tn402.5 A and Fig. Tn402.5 B) suggesting that TniQ expression and resolution are intimately coupled. Further analysis is necessary to provide direct evidence that these repeated sequences are TniR binding sites and to establish a detailed recombination mechanism.
TniR Expression and Translation Initiation
A potential promoter [11] for tniR expression is also located in this region (Fig. Tn402.5 B) and is clearly well placed to be regulated by TniR binding. Fig. Tn402.5 B also shows that the equivalent region of Tn402/Tn5090 is very similar with related repeat sequences and potential promoter elements. However, there is additional G residue and changes at the potential TniR ATG codon compared to those observed in Tn5053. However, there is a suitably placed in phase ATG codon slightly further downstream in both Tn402/Tn5090 and in Tn5053 which suggests that this maybe the true initiation codon for TniR.
TniA Expression and Translation Initiation
In addition to the ambiguity of the TniR initiation codon, a similar question arises concerning identification of the initiation codon of TniA (Fig. Tn402.6). Certain family members (e.g. Tn402 and integron members) have an orf with an initiation codon upstream of the final t repeat (Fig. Tn402.6) whereas others (e.g. Tn502 and Tn512) appear to initiate further downstream.
Alignment of the DNA sequences shows that all members are very similar in sequence in this region but that those with the downstream start codon each contain a 2 bp insertion which places the upstream region out of phase (Fig. Tn402.6). Since Tn502 and Tn512 and Tn5053 are fully functional [13][51][52], it seems possible that it is the downstream start codon that is used in all family members (Tn402.9 TniA). But this will need experimental confirmation.
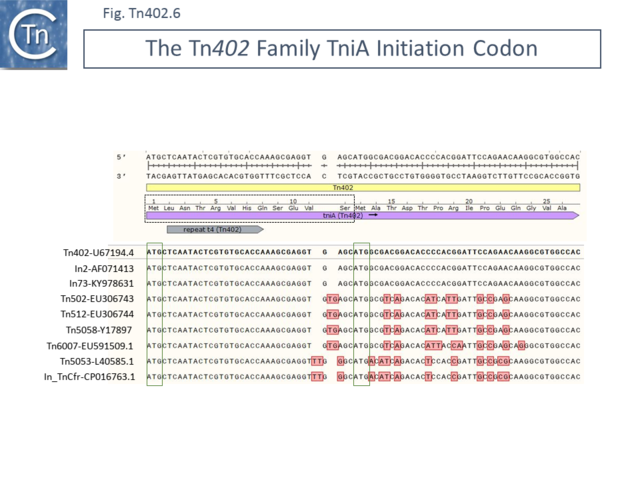
TniB Expression, Translation Initiation and Translational Coupling with TniA
A similar ambiguity arises with TniB. A DNA alignment (Fig. Tn402.7 A), however, not only shows a high degree of identity but suggests that there may be some form of translational coupling in the expression of TniA and TniB. There are potential TniB GTG start codons 1bp upstream and 2bp downstream of the predicted TniA termination codon (TAG) [53].
Although we have chosen to use the downstream GTG (Fig. Tn402.9 TniB), it should be remembered that if the upstream GTG is used, TniB would include an N-terminal extension of three amino acids, MVA. Note that the 2 mutations shown in the figure are synonymous (ATT/ATC) and conservative (GAC/GAA) respectively.
TniQ Expression, Translation Initiation and Translational Coupling with TniB
For many examples of Tn402 family transposons, the initation codon of TniQ (GTG) overlaps the termination codon (TGA) of TniB (Fig. Tn402.7 B), a situation typical of translational coupling [53]. All members of the family so far identified show high conservation and all contain the overlapping GTGA initiation/termination codons. Although some family members had included annotated TniB and or TniQ genes which had internal start codons, the most parsimonious interpretation is that the entire family follow identical initiation and termination rules (Fig. Tn402.9 TniQ).
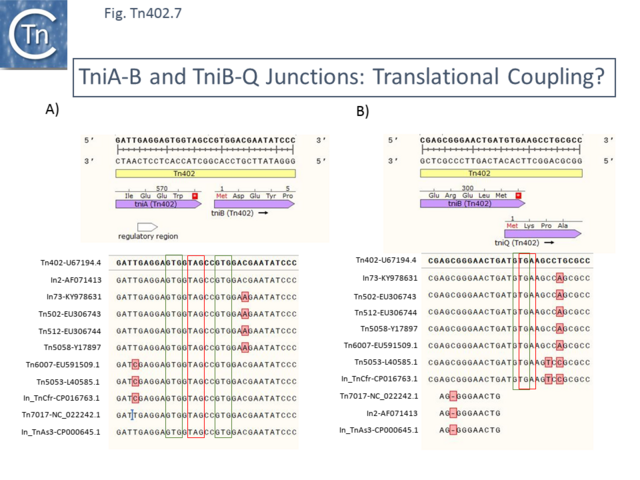
Diversity
There are three principal groups which constitute the Tn402 family based on the type of passenger genes they carry. The first, Tn402, includes those which have acquired an integron recombination platform of type 1 with an integrase IntI1 derivative. The second, TnPfu1, is a small group composed of members which have integrated a type 3 integron recombination platform with an IntI3 integrase. The third group, Tn5053, includes members which do not carry integron function but instead carry mercury resistance genes.
A phylogenetic tree of the concatenated TniA,B,Q and R proteins (Fig. Tn402.8) indicated that Tn5053 group members carrying mercury resistance genes tend to be clustered with Tn502 and Tn512 although several are intermingled with Tn402 IntI1 carrying transposons. This is perhaps not surprising since many Tn5053 members were selected using a BLAST search. A similar argument can be proposed for the three TnPfu1 related transposons which carry an IntI3 gene.
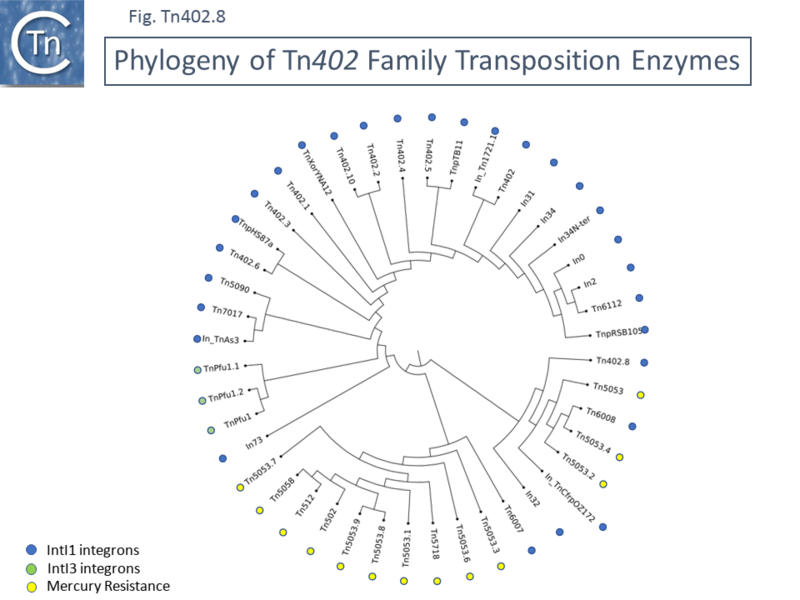
When considered individually, the different gene products are very closely related: TniA (Fig. Tn402.9 see the slide show below); TniB (Fig. Tn402.9 see the slide show below); TniQ (Fig. Tn402.9 see the slide show below) and TniR (Fig. Tn402.9 see the slide show below).
The fact that the TniA,B,Q and R proteins are highly conserved (Fig. Tn402.9) whereas the DNA sequences are more variable (see Fig. Tn402.11 and Fig. Tn402.13) (and therefore the DNA variability mainly introduces synonymous mutations together with some conservative amino acid changes; Fig. Tn402.9) suggests either that the phylogenetic branches are quite recent or that there is significant selective pressure on the protein ensemble.
The major differences between each of the Tn402 family members lie in the passenger genes which they carry.
The Tn402 group and transposon decay

As stated above, there has been some ambiguity in the literature concerning the nomenclature of Tn402 family members as transposons or as integrons and that this has been due to decay in the Tn402 tniABQR transposition module. In this group, the antibiotic resistance genes are oriented towards and downstream of the transposition genes (Fig. Tn402.1). Fig. Tn402.10 shows an alignment of an illustrative group of Tn402 elements.
All the examples include the right (integrase proximal) Tn402 end, IRi and its 19 bp repeated sequences together with the integron integrase, intI1 gene, the binding site for the host LexA protein which couples IntI1 expression to the host SOS system [8][54][55], the integron cassette promoter Pc which drives expression of the genes carried in the integron cassettes, and the attI recombination site which serves in the acquisition of integrons cassettes [8].
This is followed by the integron cassettes which vary from element to element and are indicated by the small arrow heads. Tn402 (U67194.4) itself carries three cassettes: qacH; orfD, a gene of unknown function; and the dihydrofolate reductase gene, dfrB3. The IS402 family member with the lowest number of integron cassettes is In_Tn1721.1 (HQ730118.1) which includes only qacH and its recombination sites while Tn402.5 (MH392234) [19] carries, in addition, a second cassette with a gene of unknown function.
Within this group, Tn402.5 (MH392234), In_Tn1721.1, Tn402.4 (MW150990), Tn6112,(HQ423158 ) [56], TnpHS87a (KR106190) [57], and TnXorYNA12 (HQ662557) [58] all carry a full complement of the transposition module and a variety of different integron cassettes.
A second group has lost both tniR and tniQ genes. These include: In0 (U49101) [59] where they, together with the 3’ end of tniB, have been replaced by an insertion of IS1326 upstream of a GNAT cassette; Tn7017 (NC_022242.1) [60] in which the In0 GNAT cassette abuts a further 3’ deleted tniB (presumably following excision of the IS1326 copy); TnpRSB105 (DQ839391) [29] which has a similar tniB-GNAT junction but in which a tip of IS1326 (GGCCTG) has been retained; In2 (AF071413) which contains an insertion of IS1353 into the istA gene of IS1326, a common feature of a number of integron structures; In_Tn4 (KY749247.1) [61] which, except for the absence of the IS1353 insertion, is identical to In2; In_TnAs3 (CP000645) which has an identical tniB-GNAT junction as Tn7017; In34 (AY123253 ) [62] is complex and, in addition, to carrying the IS1326::IS1353 insertion, also carries an insertion of the IS26-based transposon Tn4352 in tniA; In_Tn1935 (MK797990) is identical to In34 except for its integron cassettes; and, finally, In_Tn5045 (FN821089) [16] carries the tniB-GNAT junction and also a Tn3-family chromate resistance transposon, Tn TnOtChr.1, at the 5’ end of tniB. For In31 (AJ223604) [63], tniB is truncated further to its 5’ end than those with a tniB-GNAT junction and for In_Tn21.1 (MH257753) an IS26 insertion has led to a 3’ truncation of the tniA gene.
A third group, which includes In33 (AF313471) [64], In36 (AY259085) [65], In4 (U12338.3) [66], In37 (AY259086) [65] and In_Tn6025 (GU562437) have all undergone a complex rearrangement involving IS6100 and duplication of IRt and repeats in an inverted orientation surrounding the IS6100 copy.
The fourth group, including In7 (L06418)[67], In6_p (L06822) and InS21 are closely related but are only partially sequenced examples. They do not include an IRt end and include an insertion of ISCR1 (IS91 and ISCR family). However, the reported partial integron In7 (KU997026) from Escherichia coli plasmid pDGO100 [68] closely resembles In34. It extends both upstream to include the first IS26 copy of transposon Tn4352 inserted in tniA of In34 and downstream to include the IRi end. The In7 (L06418) copy [67] is a sub-sequence of this. A similar problem is likely to arise with In6_p (L06822) [69] which also shares several identical regions with In34. In6_p includes a full set of IRi sequences downstream of its IntI1 gene but the sequence of its pSa plasmid parent [70] is not available. Finally, InS21 (AJ311891), again with significant regions identical to In34, from plasmid pS21, carries a complete IRi end but is truncated at the opposite end.
There is consequently no strong evidence that integron platforms along with their cassettes have been transferred without their accompanying Tn402 transposition system. “Integron” insertion may have occurred prior to decay of the tniABQR genes (see [21]).
Inter-transposon recombination at the Tn402 res
As in other transposons which use a site-specific resolution mechanism as a step in their transposition [71][72], members of the IS402 family can also undergo inter-transposon recombination at the res site and in this way exchange DNA modules. This type of behavior has been reported by Labbate et al. [72][73] who observed that transposon Tn6007 from Enterobacter cloacae included a class 1 integron with two non-antibiotic-resistance-type gene cassettes and a complete transposition module. The tniABQR module appeared to be a hybrid with a boundary within the res site (Fig. Tn402.11). The level of identity from the res site through tniR and the integron was observed to be high (99.9%) while that further upstream through the tniA,B and Q genes was only 89% [73]. This is not an isolated case and a number of additional examples of this phenomenon can also be observed (Fig. Tn402.11). Similar observations were made by Betteridge et al. in an analysis of integron platforms isolated from Human Commensal Bacteria in an Intensive Care Unit (ICU) [74].
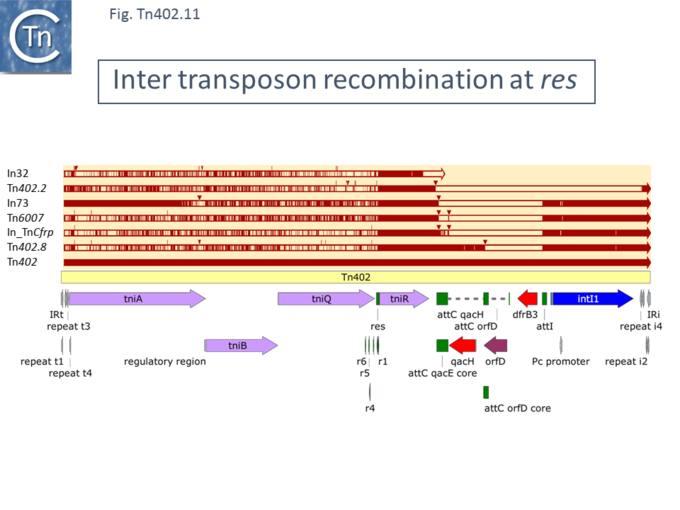
Origin of the Tn402 group integrons.
One hypothesis to explain the origin of the mobilizable class 1 integron is that it is derived from a chromosomal integron platform that became mobile by the acquisition of Tn402-like transposition functions. However, the identification of two other Tn402 family groups with totally different passenger gene ensembles tends to suggest that it might have been an ancestral Tn402 transposon which acquired the passengers.
The suggestion that some class 1 integrons identified in a sediment microbial community potentially predate integron association with Tn402 is difficult to assess [21]. Also, no ancestral Tn402 transposon has yet been identified without passenger genes.
The TnPfu1 group
In the majority of Tn402 family transposons, integron platform is of type 1 and the integrase gene is known as intI1. IntI1 is invariably located at the distal transposon end and is expressed in the same direction as tniA, however, there is an additional group in which the integrase faces towards tniR.
An example of a Tn402 derivative with this configuration of tniA and intI genes, TnPfu1 (LC331665.1), (Fig. Tn402.11) was first identified from Pseudomonas fulva and carried a blaIMP-1 gene together with AAC(6') and fosX gene cassettes [12]. A BLAST search for tniA -intI1 regions with this configuration revealed only two additional examples, all closely related except for the gene cassettes carried by the integron platform: TnPfu1.1 (LC589064) carries a bla GES gene and a cassette containing AAC(6')-Ib8/-Ib4, while TnPfu1.2 (LC589062) includes two copies of bla GES.
Members of this group carry another type of intI gene associated with Tn402-like transposition genes, intI3. This was first identified in Serratia marcescens plasmid pSMB731 as part of a new integron type, type 3 [75], in which the integrase showed only limited identity to IntI1. Unfortunately, only partial sequence data was presented (Fig. Tn402.12) although a limited extension was published more recently [76] covering the tniA gene upstream and an AAC(6')-Ib8 gene cassette downstream together with the IRi end (Fig. Tn402.12).
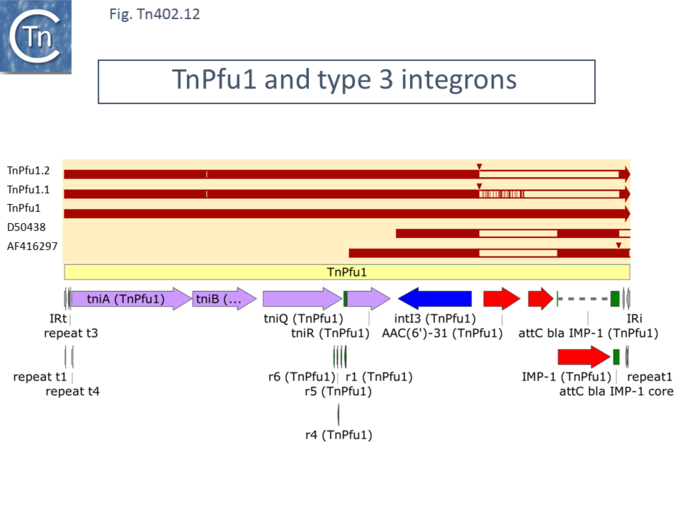
The Tn5053 group
Tn5053 (L40585.1) itself (Fig. Tn402.4), isolated from a Russian mercury mine [13], carries an operon specifying resistance to mercury. The mercury resistance genes, like the antibiotic resistance genes in the Tn402 group, are oriented such that expression occurs towards and downstream of the transposition genes. For the sake of simplicity, the IR distal to the transposition genes has retained its name from Tn402, IRi.
As for the Tn402 and TnPfu1 groups, the TniA,B,Q and R proteins are well conserved (Fig. Tn402.9) while the DNA sequences show a higher degree of variation (Fig. Tn402.13).
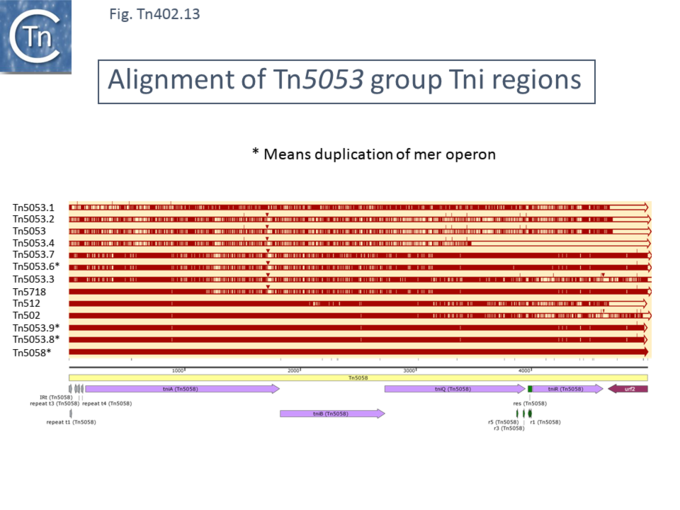
Inter-transposon recombination at the Tn5053 res
Inter-transposon recombination at the res site is also observed in the case of family members which carry mercury resistance genes in place of the integron platform. This can be seen between Tn5053 (L40585.1) and a related mercury resistance transposon we have called Tn5053.1 (CP002451) from the Alicycliphilus denitrificans BC plasmid pALIDE02 [77] (Fig. Tn402.14) where the tniABQ module has clearly been exchanged . This is would at least partially explain why Tn5053 and Tn5053.1 are located in different subgroups in the phylogenetic tree (Fig. Tn402.8)
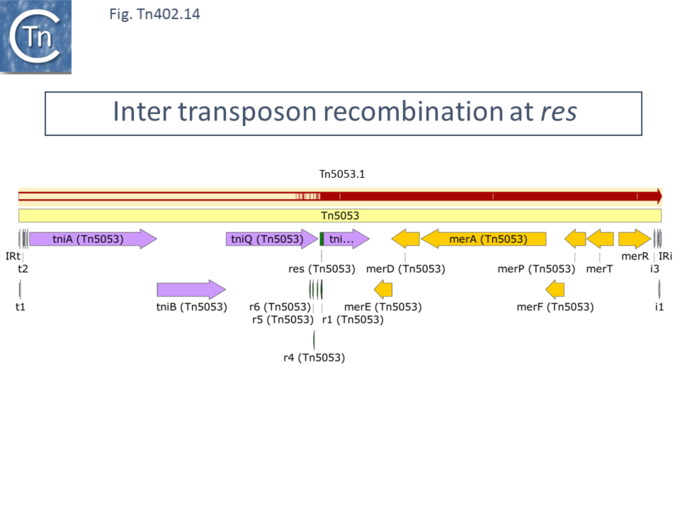
Presence of a Tn21-like IR
The relationship between members of this mercury resistance Tn402 group is complex. One close relative, Tn5053.4 (CP001919), carries a simple insertion of IS4321 at the distal end.
Interestingly, all mercury resistance Tn402 family members carry a sequence resembling a Tn21 IR, suggesting that the mercury resistance genes may have be acquired from a Tn21-derivative transposon towards the IRi end (Fig. Tn402.15) , suggesting that mercury resistance was originally acquired from a Tn21-related Tn3 family transposon (see [78]). However, Tn5053.2 (CP024682) does not and is also missing the MerA sequence up to and including merR due to insertion of an ahp gene Moreover, in a subgroup, there has been a partial duplication of the mercury resistance operon followed by some decay and rearrangement with changes in gene number and order. This may have involved the Tn21-like IR since these transposons, Tn5053.6 (JX469833), Tn5053.8 (CP017991), Tn5053.9 (CP048650), Tn5058 (Y17897) and Tn50580 (AM048832), all carry two Tn21-like IR.
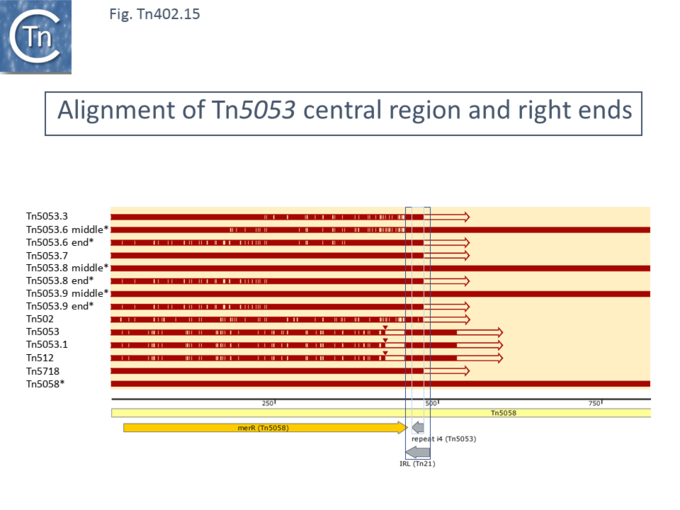
Tn5053 group Mercury resistance genes
Mercury resistance genes are generally grouped into an operon. The major players are: merA, encoding a mercuric reductase (MerA), which catalyses reduction of Hg(II) to volatile Hg(0) and is the central enzyme in Hg(II) resistance [79][80]; merB encoding an organomercury lyase (MerB); merP encoding a periplasmic Hg(II) scavenging protein (MerP); merT, merC, merE, merF and merG which encode inner membrane spanning proteins (MerT, MerC, MerE, MerF, MerG) that transport Hg(II) to the cytoplasm where it is reduced by MerA; merR and merD encoding the regulatory proteins (MerR, MerD). MerR is a transcriptional repressor in the absence of Hg(II) and an activator in its presence and MerD downregulates the operon (for review see [81]) .
Mercury resistance operons do not all carry a complete set of these genes. Fig. Tn402.16 shows a sample of mercury resistance operons from various transposons of the Tn3 and Tn402 families together with the ensemble of mer genes in the order in which they occur in the canonical Tn3 family transposon Tn21.
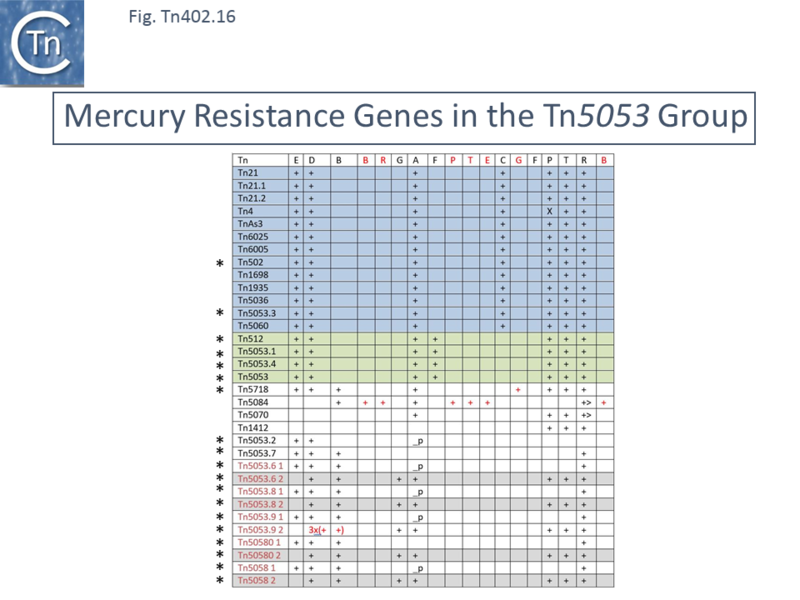
Insertion Sites: res Hunters
In early studies describing Tn502 and its differences with other mercury resistance transposons known at the time [82] it was observed that it displays a preference for insertion into a single PstI fragment of the plasmid RP1 [51]. Further restriction mapping led to the conclusion that the insertions occurred at the same site, largely in one orientation. Deletion of this region resulted in a marked reduction in Tn502 insertion frequency into the plasmid [82]. This region was subsequently shown to carry the non-transcribed spacer of the par locus of plasmid RP1, initially defined in the related plasmids RK2 and RP4 as a region required for plasmid stabilisation [83][84]. It is similarly organised to the res region of Tn3 family transposon (see [71][78]) and contains three binding sites for the ParA site-specific recombinase (resI, resII and resIII ) [85].
A similar sequence-specific insertion of Tn402/Tn5090 into derivatives of plasmids R388 and RP1 was also demonstrated experimentally [86]. In RP1, insertions occurred in two major clusters (Fig. Tn402.17 A) within 140 bp: one cluster (1) was located in the resII resolvase subsite and the second (2) within the parC gene. The orientation of Tn402 insertions in each cluster was inverted. A single major insertion site was observed in plasmid R388, again, close to the proposed res sites [86]. Furthermore, this targeting required an intact ParA since partial deletion significantly reduced the attractiveness of the derivative plasmid as a target. Moreover, a plasmid in which the ParA active site serine was mutated completely retained its activity as a transposition target [86] showing that it is the binding of resolvase and presumably the architecture of the res-ParA nucleoprotein complex rather than recombination activity which determine target activity.
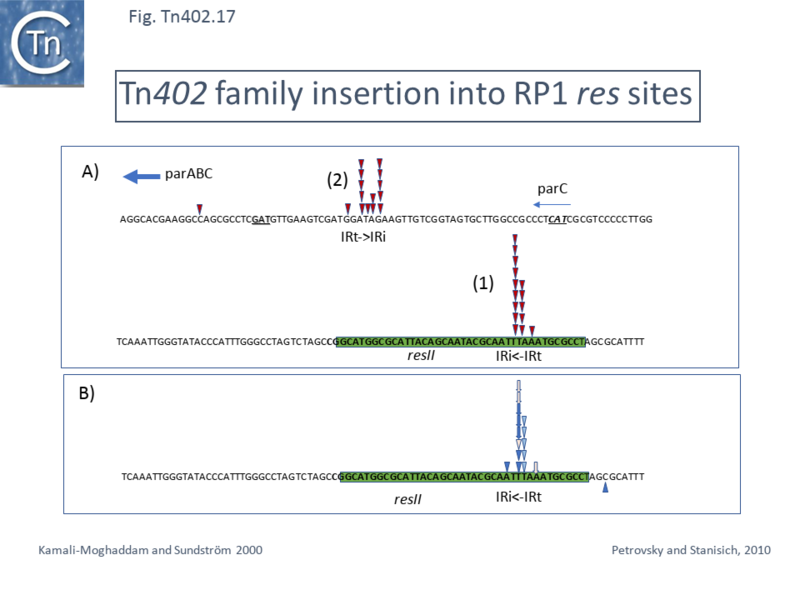
Tn2521, a multi-resistant transposon identified in the chromosome of Pseudomonas aeruginosa clinical isolates in 1982 [87] was also observed to have marked integration site-specificity in transposition into IncP plasmids (in this case R18-1, identical to R68). Tn2521 was later shown to be an integron (In33) without tni genes and its insertion point in the R18-18::Tn2521 example examined was shown to have inserted at the R18-18 equivalent to RP1/RP4 site resII [64]. Moreover, additional analysis of the flanking sequences of other integron structures indicated that insertion occurred in the res sites of several Tn3 family transposon [88]: In28 in the resI region of Tn1403; In4 abutting resI in Tn1696; In1 upstream of resP in R46 ( [89], also called TP120 [90]), the region expected to include the plasmid res site which has an adjacent functional resolvase-like gene, uvp1 [91], and is similar to and can undergo recombination with that of Tn1 [92][93][94]. In these examples, the inserted integron platform does not contain the set of Tn402 transposition enzymes. This implies either that transposition preceded decay of the transposition genes or that their products were provided in trans from another transposon in the cell as is suggested by transposition of In33 (Tn2521). A further example, however, is the insertion into Tn1721 of an integron with a full complement of Tn402 transposition enzymes (HQ730118) [74].
This type of insertion site-specificity was also observed for Tn5053 [13], again using RP1 and an RP4 derivative plasmid target. Sequence analysis showed that the insertions were flanked by a 5 bp direct target repeat as expected [13][24]. A survey of conjugative plasmids (cited in [95]) identified only a single example which acted as an efficient Tn5053 target. This plasmid carried Tn701, a close relative of Tn1721 and The Tn5053 insertion were reported to have occurred in the res site of this transposon. Introduction of Tn701 or Tn1721 into plasmids not generally active targets such as R387, pOXGen or pBR322, these became efficient Tn5053 targets [95]. In addition, a database search using the first 50 bp of the ends of Tn5053 and Tn402 as the query sequences revealed a number of Tn junctions in which the IRi ends were located upstream of resolvase-like genes [95].
In these studies [95], Insertion of Tn5035 was observed experimentally using as target the cloned insertion-free res site of Tn5044 [96] together with the native TnpR gene. Very strong clustering was observed between resI and resII and all but one insertion occurred in the same orientation (Fig. Tn402.17 C). Moreover, when cloned and in the absence of TnpR, the res sites of Tn1721 [97], Tn5036 [18], Tn5051 (no sequence available) [18][95] and Tn5059 (Y10102, partial sequence) were not attractive to Tn5035 insertion, however, when TnpR from Tn1721 was supplied in trans, efficient targeted insertion occurred and sequence analysis showed strong clustering (Fig. Tn402.17 D). This behavior rules out the possibility that expression (transcription or translation) of TnpR in its natural proximity to res provides a suitable integration substrate. A similar requirement for the ParA resolvase was reported for insertion into the RP1 res. Surprisingly target attractiveness appeared to be limited to the Tn21 branch of the Tn3 family [6][98] neither Tn1 nor Tn1000 were found to be efficient targets for Tn5053 insertion.
Tn402 family members with mercury resistance genes, Tn502 and Tn512, have also been found to show quasi-identical insertion specificity in an RP1 res target [52] as Tn402 itself (Fig. Tn402.17 B). Remarkably, the insertion site observed in resII was identical to that observed for Tn402/Tn5090 (cluster 1 in Fig. Tn402.17 A)[86]. The authors also address the potential mobility of integron platforms, such as In0 (U49101) and In2 (AF071413) which do not contain a full set of Tn402 tni genes but carry both IRi and IRt ends and are found with flanking 5 bp direct target repeats. Transposition of In2 from its location in Tn21 and of In0 from its location in plasmid pVS1 was tested by supplying Tn502 tni genes in trans. Transposition occurred at significant frequencies if both tni and the RP1 par site were present. Moreover, the vast majority of insertions occurred in the expected orientation at the cluster 1 site (Fig. Tn402.17 B). One puzzling observation was that although neither In0 nor In2 include a Tn402 res site never-the-less, the transposition products had undergone resolution.
Complementation by a resident Tn402 family transposon would provide an explanation for transposition of Tn2521 (In33) into the R18-1 [87] res site even though it does not contain a single Tn402 transposition gene [64].
In summary, all members of the Tn402 family tested appear to target res sites of plasmids and Tn3 family transposons of the Tn21 group. They require the cognate resolvase protein [86][95] but do not require it to be catalytically active [86]. The role of TnpR could be structural, providing an architecturally precise nucleoprotein complex for integration and/or furnishing an interface for direct protein-protein interaction with a TniABQ component. Transposons with tni gene sets can also complement in trans integron platforms which are lacking these genes [52] in principal providing the capacity to mobilise non-autonomous integron structures.
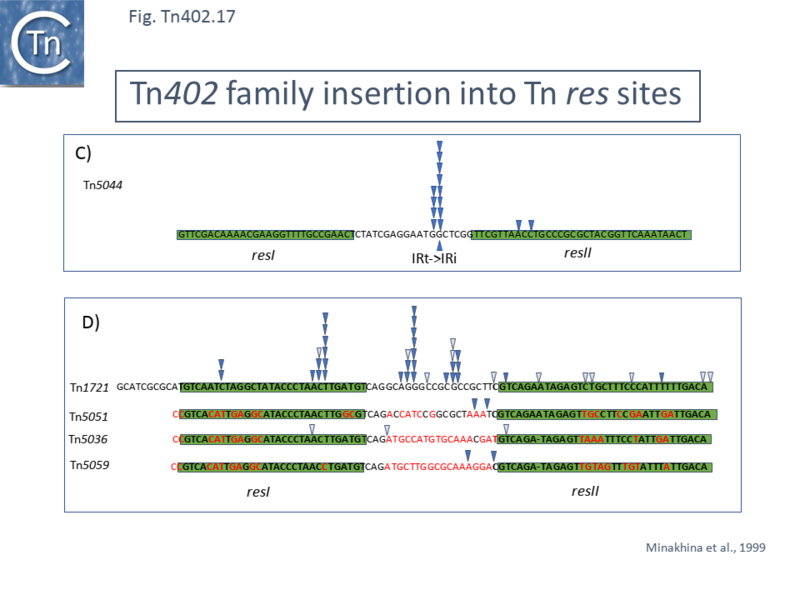
A reflection on transposition mechanism.
Except for the bioinformatic analysis of their transposition proteins and the demonstration that their transposition requires a cointegration step which is resolved at a specific site, the res site and involves the TniR site-specific recombinase [11], there has been no significant biochemical or structural studies of Tn402 family transposition.
Studies with purified TniA [36] have shown that the protein binds to the left IRt end of Tn402/Tn5090 to generate six retarded bands (I-VI). This was further explored using DNase footprinting which showed that the four sites t1-t4 were Species I-III or of the resolvase TniR to the repeated sequences at the res site. It is therefore not known which of the multiple end sites are important for transposition or whether all repeated sequences in and around the proposed crossover point between r1 and r2 are necessary for resolution.
Neither have the roles played by TniA, TniB or TniQ been determined. The fact that TniA carries a DDE motif and shows 25% similarity to TnsB transposase [99] of transposon Tn7 [10][11] strongly suggests that it is the Tn402 family transposase and the observation that TniB shares similarities with TnsC suggests that it may play a DNA-dependent bridging reaction between TniA and TniQ. It seems possible that, like TnsD and TnsE of Tn7 [99][100][101], tniQ is involved in the choice of the target sequence. Binding to the small repeated sequences at IRt and IRi also has yet to be demonstrated.
In view of the sequence similarities between the Tn7 and Tn402 family transposition proteins, it is interesting to note that the Tn402 family possesses a dedicated system to resolve cointegrates whereas Tn7 does not. In contrast, Tn7 encodes two endonucleases, TnsA and TnsB. The DDE transposase, TnsB, binds to the Tn7 ends cleaves only one strand of the transposon whereas TnsA, which resembles a restriction endonuclease [102], cleaves the complementary strand. Together, both, form a heteromeric complex and in concert, achieve cleavage and joining reactions at both Tn7 ends [103]. Transposition of Tn7 normally occurs by a cut-and-paste mechanism [104]. However, although TnsA is required for TnsB activity, mutation of the active site of TnsA, prevents the cut-and-paste pathway and results in the formation of cointegrates [105]. Therefore, it seems probable that TniA, like TnsB, catalyses only single-strand cleavage at each transposon end and to deal with the resulting cointegrate structures, Tn402 has recruited a resolvase-like protein and an accompanying abutting res site while Tn7 has recruited a second restriction endonuclease protein.
An additional observation is that the majority of Tn402-family transposons and integrons are flanked by 5 bp direct target repeats. This implies that intermolecular transposition represents a significant, if not unique pathway since intramolecular transposition using a cointegrate pathway leads to loss of the direct target repeats (Fig.17.1 and Fig.17.2)
Acknowledgements
We would like to thank Steve Petrovski (La Trobe University, Australia) for critical comments.
Bibliography
- ↑ 1.0 1.1 Shapiro & Sporn. Tn402: a new transposable element determining trimethoprim resistance that inserts in bacteriophage lambda. Journal of bacteriology. 1977. 129. pp. 1632-5. doi: 10.1128/jb.129.3.1632-1635.1977. PMID: 321437.
- ↑ Jobanputra & Datta. Trimethoprim R factors in enterobacteria from clinical specimens. Journal of medical microbiology. 1974. 7. pp. 169-77. doi: 10.1099/00222615-7-2-169. PMID: 4599661.
- ↑ Jacob A, Shapiro J, Yamamoto L, Smith DI, Cohen SN, Berg D. . Plasmids studied in Escherichia coli and other enteric bacteria. In (ed.),. In: Bukhari AI, Shapiro J, Adhya S, editors. DNA insertion elements, episomes and plasmids . Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.; 1977.
- ↑ Stokes & Hall. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Molecular microbiology. 1989. 3. pp. 1669-83. doi: 10.1111/j.1365-2958.1989.tb00153.x. PMID: 2560119.
- ↑ Brown et al.. The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Molecular & general genetics : MGG. 1986. 202. pp. 143-51. doi: 10.1007/BF00330531. PMID: 3007931.
- ↑ 6.0 6.1 Grinsted et al.. The Tn21 subgroup of bacterial transposable elements. Plasmid. 1990. 24. pp. 163-89. doi: 10.1016/0147-619x(90)90001-s. PMID: 1963947.
- ↑ Brown et al.. The integrons In0, In2, and In5 are defective transposon derivatives. Journal of bacteriology. 1996. 178. pp. 4429-37. doi: 10.1128/jb.178.15.4429-4437.1996. PMID: 8755869.
- ↑ 8.0 8.1 8.2 Escudero et al.. The Integron: Adaptation On Demand. Microbiology spectrum. 2015. 3. pp. MDNA3-0019-2014. doi: 10.1128/microbiolspec.MDNA3-0019-2014. PMID: 26104695.
- ↑ Mazel. Integrons: agents of bacterial evolution. Nature reviews. Microbiology. 2006. 4. pp. 608-20. doi: 10.1038/nrmicro1462. PMID: 16845431.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 10.6 10.7 10.8 Rådström et al.. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. Journal of bacteriology. 1994. 176. pp. 3257-68. doi: 10.1128/jb.176.11.3257-3268.1994. PMID: 8195081.
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 11.6 11.7 Kholodii et al.. Four genes, two ends, and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Molecular microbiology. 1995. 17. pp. 1189-200. doi: 10.1111/j.1365-2958.1995.mmi_17061189.x. PMID: 8594337.
- ↑ 12.0 12.1 Yamamoto et al.. Molecular Analysis of a blaIMP-1-Harboring Class 3 Integron in Multidrug-Resistant Pseudomonas fulva. Antimicrobial agents and chemotherapy. 2018. 62. doi: 10.1128/AAC.00701-18. PMID: 29784850.
- ↑ 13.0 13.1 13.2 13.3 13.4 13.5 13.6 13.7 Kholodii et al.. Tn5053, a mercury resistance transposon with integron's ends. Journal of molecular biology. 1993. 230. pp. 1103-7. doi: 10.1006/jmbi.1993.1228. PMID: 8387603.
- ↑ 14.0 14.1 Jobling MG, Peters SE, Ritchie DA. Plasmid-borne mercury resistance in aquatic bacteria. FEMS Microbiol Lett. 1988;49:31–37.
- ↑ 15.0 15.1 Mindlin et al.. Present-day mercury resistance transposons are common in bacteria preserved in permafrost grounds since the Upper Pleistocene. Research in microbiology. 2005. 156. pp. 994-1004. doi: 10.1016/j.resmic.2005.05.011. PMID: 16084067.
- ↑ 16.0 16.1 Petrova et al.. Tn5045, a novel integron-containing antibiotic and chromate resistance transposon isolated from a permafrost bacterium. Research in microbiology. 2011. 162. pp. 337-45. doi: 10.1016/j.resmic.2011.01.003. PMID: 21262357.
- ↑ Vishnivetskaya & Kathariou. Putative transposases conserved in Exiguobacterium isolates from ancient Siberian permafrost and from contemporary surface habitats. Applied and environmental microbiology. 2005. 71. pp. 6954-62. doi: 10.1128/AEM.71.11.6954-6962.2005. PMID: 16269730.
- ↑ 18.0 18.1 18.2 Yurieva et al.. Intercontinental spread of promiscuous mercury-resistance transposons in environmental bacteria. Molecular microbiology. 1997. 24. pp. 321-9. doi: 10.1046/j.1365-2958.1997.3261688.x. PMID: 9159519.
- ↑ 19.0 19.1 19.2 Shintani et al.. Plant Species-Dependent Increased Abundance and Diversity of IncP-1 Plasmids in the Rhizosphere: New Insights Into Their Role and Ecology. Frontiers in microbiology. 2020. 11. pp. 590776. doi: 10.3389/fmicb.2020.590776. PMID: 33329469.
- ↑ 20.0 20.1 Schneiker et al.. The genetic organization and evolution of the broad host range mercury resistance plasmid pSB102 isolated from a microbial population residing in the rhizosphere of alfalfa. Nucleic acids research. 2001. 29. pp. 5169-81. doi: 10.1093/nar/29.24.5169. PMID: 11812851.
- ↑ 21.0 21.1 21.2 Stokes et al.. Class 1 integrons potentially predating the association with tn402-like transposition genes are present in a sediment microbial community. Journal of bacteriology. 2006. 188. pp. 5722-30. doi: 10.1128/JB.01950-05. PMID: 16885440.
- ↑ 22.0 22.1 22.2 Smalla et al.. Increased abundance of IncP-1beta plasmids and mercury resistance genes in mercury-polluted river sediments: first discovery of IncP-1beta plasmids with a complex mer transposon as the sole accessory element. Applied and environmental microbiology. 2006. 72. pp. 7253-9. doi: 10.1128/AEM.00922-06. PMID: 16980416.
- ↑ Hobman & Crossman. Bacterial antimicrobial metal ion resistance. Journal of medical microbiology. 2015. 64. pp. 471-497. doi: 10.1099/jmm.0.023036-0. PMID: 25418738.
- ↑ 24.0 24.1 Hobman J, Kholodii G, Nikiforov V, Ritchie DA, Strike P, Yurieva O. The sequence of the mer operon of pMER327/419 and transposon ends of pMER327/419, 330 and 05. Gene. 1994 Aug;146(1):73–78.
- ↑ Essa et al.. Mercury resistance determinants related to Tn21, Tn1696, and Tn5053 in enterobacteria from the preantibiotic era. Antimicrobial agents and chemotherapy. 2003. 47. pp. 1115-9. doi: 10.1128/AAC.47.3.1115-1119.2003. PMID: 12604550.
- ↑ Datta & Hughes. Plasmids of the same Inc groups in Enterobacteria before and after the medical use of antibiotics. Nature. 1983. 306. pp. 616-7. doi: 10.1038/306616a0. PMID: 6316165.
- ↑ Martinez et al.. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. Journal of bacteriology. 2001. 183. pp. 5684-97. doi: 10.1128/JB.183.19.5684-5697.2001. PMID: 11544232.
- ↑ Mindlin et al.. Mercury resistance transposons of gram-negative environmental bacteria and their classification. Research in microbiology. 2001. 152. pp. 811-22. doi: 10.1016/s0923-2508(01)01265-7. PMID: 11763242.
- ↑ 29.0 29.1 Schlüter et al.. Erythromycin resistance-conferring plasmid pRSB105, isolated from a sewage treatment plant, harbors a new macrolide resistance determinant, an integron-containing Tn402-like element, and a large region of unknown function. Applied and environmental microbiology. 2007. 73. pp. 1952-60. doi: 10.1128/AEM.02159-06. PMID: 17261525.
- ↑ Szczepanowski et al.. The 120 592 bp IncF plasmid pRSB107 isolated from a sewage-treatment plant encodes nine different antibiotic-resistance determinants, two iron-acquisition systems and other putative virulence-associated functions. Microbiology (Reading, England). 2005. 151. pp. 1095-1111. doi: 10.1099/mic.0.27773-0. PMID: 15817778.
- ↑ Tennstedt et al.. Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS microbiology ecology. 2003. 45. pp. 239-52. doi: 10.1016/S0168-6496(03)00164-8. PMID: 19719593.
- ↑ Wibberg et al.. The IncF plasmid pRSB225 isolated from a municipal wastewater treatment plant's on-site preflooder combining antibiotic resistance and putative virulence functions is highly related to virulence plasmids identified in pathogenic E. coli isolates. Plasmid. 2013. 69. pp. 127-37. doi: 10.1016/j.plasmid.2012.11.001. PMID: 23212116.
- ↑ Oliveira et al.. Comparative genomics of IncP-1ε plasmids from water environments reveals diverse and unique accessory genetic elements. Plasmid. 2013. 70. pp. 412-9. doi: 10.1016/j.plasmid.2013.06.002. PMID: 23831558.
- ↑ Liebert et al.. Transposon Tn21, flagship of the floating genome. Microbiology and molecular biology reviews : MMBR. 1999. 63. pp. 507-22. doi: 10.1128/MMBR.63.3.507-522.1999. PMID: 10477306.
- ↑ Sundström et al.. Characterization of transposon Tn5086, carrying the site-specifically inserted gene dhfrVII mediating trimethoprim resistance. Journal of bacteriology. 1993. 175. pp. 1796-805. doi: 10.1128/jb.175.6.1796-1805.1993. PMID: 8383666.
- ↑ 36.0 36.1 36.2 36.3 Kamali-Moghaddam & Sundström. Arrayed transposase-binding sequences on the ends of transposon Tn5090/Tn402. Nucleic acids research. 2001. 29. pp. 1005-11. doi: 10.1093/nar/29.4.1005. PMID: 11160934.
- ↑ Lichtenstein C, Brenner S. Site-Specific Properties of Tn7 Transposition into the E. coil Chromosome. Mol Gen Genet. 1981;183:380–387.
- ↑ McKown RL, Waddell CS, Arciszewska LK, Craig NL. Identification of a transposon Tn7-dependent DNA-binding activity that recognizes the ends of Tn7. ProcNatlAcadSciUSA. 1987;84:7807–7811.
- ↑ Arciszewska & Craig. Interaction of the Tn7-encoded transposition protein TnsB with the ends of the transposon. Nucleic acids research. 1991. 19. pp. 5021-9. doi: 10.1093/nar/19.18.5021. PMID: 1656385.
- ↑ Craigie et al.. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984. 39. pp. 387-94. doi: 10.1016/0092-8674(84)90017-5. PMID: 6094016.
- ↑ Montaño et al.. The μ transpososome structure sheds light on DDE recombinase evolution. Nature. 2012. 491. pp. 413-7. doi: 10.1038/nature11602. PMID: 23135398.
- ↑ Mahillon & Chandler. Insertion sequences. Microbiology and molecular biology reviews : MMBR. 1998. 62. pp. 725-74. doi: 10.1128/MMBR.62.3.725-774.1998. PMID: 9729608.
- ↑ Arciszewska & Craig. Interaction of the Tn7-encoded transposition protein TnsB with the ends of the transposon. Nucleic acids research. 1991. 19. pp. 5021-9. doi: 10.1093/nar/19.18.5021. PMID: 1656385.
- ↑ Craigie et al.. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984. 39. pp. 387-94. doi: 10.1016/0092-8674(84)90017-5. PMID: 6094016.
- ↑ Stellwagen & Craig. Gain-of-function mutations in TnsC, an ATP-dependent transposition protein that activates the bacterial transposon Tn7. Genetics. 1997. 145. pp. 573-85. doi: 10.1093/genetics/145.3.573. PMID: 9055068.
- ↑ Gamas & Craig. Purification and characterization of TnsC, a Tn7 transposition protein that binds ATP and DNA. Nucleic acids research. 1992. 20. pp. 2525-32. doi: 10.1093/nar/20.10.2525. PMID: 1317955.
- ↑ Choi et al.. The Tn7 transposition regulator TnsC interacts with the transposase subunit TnsB and target selector TnsD. Proceedings of the National Academy of Sciences of the United States of America. 2014. 111. pp. E2858-65. doi: 10.1073/pnas.1409869111. PMID: 24982178.
- ↑ Maxwell et al.. B protein of bacteriophage mu is an ATPase that preferentially stimulates intermolecular DNA strand transfer. Proceedings of the National Academy of Sciences of the United States of America. 1987. 84. pp. 699-703. doi: 10.1073/pnas.84.3.699. PMID: 2949325.
- ↑ Plasterk RH, Brinkman A, van de Putte P. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. ProcNatlAcadSciUSA. 1983;80:5355–5358.
- ↑ 50.0 50.1 Galas DJ, Chandler M. Structure and stability of Tn9-mediated cointegrates. J Mol Biol. 1982 Jan;154(2):245–272.
- ↑ 51.0 51.1 Grinsted et al.. Properties of a R factor which originated in Pseudomonas aeruginosa 1822. Journal of bacteriology. 1972. 110. pp. 529-37. doi: 10.1128/jb.110.2.529-537.1972. PMID: 4336689.
- ↑ 52.0 52.1 52.2 Petrovski & Stanisich. Tn502 and Tn512 are res site hunters that provide evidence of resolvase-independent transposition to random sites. Journal of bacteriology. 2010. 192. pp. 1865-74. doi: 10.1128/JB.01322-09. PMID: 20118251.
- ↑ 53.0 53.1 Huber et al.. Translational coupling via termination-reinitiation in archaea and bacteria. Nature communications. 2019. 10. pp. 4006. doi: 10.1038/s41467-019-11999-9. PMID: 31488843.
- ↑ Baharoglu et al.. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS genetics. 2010. 6. pp. e1001165. doi: 10.1371/journal.pgen.1001165. PMID: 20975940.
- ↑ Cambray et al.. Integrons. Annual review of genetics. 2010. 44. pp. 141-66. doi: 10.1146/annurev-genet-102209-163504. PMID: 20707672.
- ↑ Sajjad et al.. Preclinical class 1 integron with a complete Tn402-like transposition module. Applied and environmental microbiology. 2011. 77. pp. 335-7. doi: 10.1128/AEM.02142-10. PMID: 21037292.
- ↑ Bi et al.. A Site-Specific Integrative Plasmid Found in Pseudomonas aeruginosa Clinical Isolate HS87 along with A Plasmid Carrying an Aminoglycoside-Resistant Gene. PloS one. 2016. 11. pp. e0148367. doi: 10.1371/journal.pone.0148367. PMID: 26841043.
- ↑ Xu et al.. Identification and characterization of integron-mediated antibiotic resistance in the phytopathogen Xanthomonas oryzae pv. oryzae. PloS one. 2013. 8. pp. e55962. doi: 10.1371/journal.pone.0055962. PMID: 23437082.
- ↑ Bissonnette & Roy. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. Journal of bacteriology. 1992. 174. pp. 1248-57. doi: 10.1128/jb.174.4.1248-1257.1992. PMID: 1310501.
- ↑ Di Pilato et al.. Characterization of plasmid pAX22, encoding VIM-1 metallo-β-lactamase, reveals a new putative mechanism of In70 integron mobilization. The Journal of antimicrobial chemotherapy. 2014. 69. pp. 67-71. doi: 10.1093/jac/dkt311. PMID: 23928025.
- ↑ Cox & Schildbach. Sequence of the R1 plasmid and comparison to F and R100. Plasmid. 2017. 91. pp. 53-60. doi: 10.1016/j.plasmid.2017.03.007. PMID: 28359666.
- ↑ Partridge & Hall. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrobial agents and chemotherapy. 2003. 47. pp. 342-9. doi: 10.1128/AAC.47.1.342-349.2003. PMID: 12499211.
- ↑ Laraki et al.. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrobial agents and chemotherapy. 1999. 43. pp. 890-901. doi: 10.1128/AAC.43.4.890. PMID: 10103196.
- ↑ 64.0 64.1 64.2 Partridge et al.. Characterization and movement of the class 1 integron known as Tn2521 and Tn1405. Antimicrobial agents and chemotherapy. 2002. 46. pp. 1288-94. doi: 10.1128/AAC.46.5.1288-1294.2002. PMID: 11959558.
- ↑ 65.0 65.1 Wang et al.. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrobial agents and chemotherapy. 2003. 47. pp. 2242-8. doi: 10.1128/AAC.47.7.2242-2248.2003. PMID: 12821475.
- ↑ Bissonnette et al.. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. Journal of bacteriology. 1991. 173. pp. 4493-502. doi: 10.1128/jb.173.14.4493-4502.1991. PMID: 1648560.
- ↑ 67.0 67.1 Cameron et al.. Nucleotide sequence of the AAD(2) aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic acids research. 1986. 14. pp. 8625-35. doi: 10.1093/nar/14.21.8625. PMID: 3024112.
- ↑ Harmer et al.. pDGO100, a type 1 IncC plasmid from 1981 carrying ARI-A and a Tn1696-like transposon in a novel integrating element. Plasmid. 2016. 86. pp. 38-45. doi: 10.1016/j.plasmid.2016.06.002. PMID: 27318267.
- ↑ Stokes et al.. The partial 3'-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid. 1993. 30. pp. 39-50. doi: 10.1006/plas.1993.1032. PMID: 8378445.
- ↑ Ireland. Detailed restriction enzyme map of crown gall-suppressive IncW plasmid pSa, showing ends of deletion causing chloramphenicol sensitivity. Journal of bacteriology. 1983. 155. pp. 722-7. doi: 10.1128/jb.155.2.722-727.1983. PMID: 6307978.
- ↑ 71.0 71.1 Lima-Mendez et al.. Toxin-Antitoxin Gene Pairs Found in Tn3 Family Transposons Appear To Be an Integral Part of the Transposition Module. mBio. 2020. 11. doi: 10.1128/mBio.00452-20. PMID: 32234815.
- ↑ 72.0 72.1 Yano et al.. Cointegrate-resolution of toluene-catabolic transposon Tn4651: determination of crossover site and the segment required for full resolution activity. Plasmid. 2013. 69. pp. 24-35. doi: 10.1016/j.plasmid.2012.07.004. PMID: 22878084.
- ↑ 73.0 73.1 Labbate et al.. A class 1 integron present in a human commensal has a hybrid transposition module compared to Tn402: evidence of interaction with mobile DNA from natural environments. Journal of bacteriology. 2008. 190. pp. 5318-27. doi: 10.1128/JB.00199-08. PMID: 18502858.
- ↑ 74.0 74.1 Betteridge et al.. Genetic context and structural diversity of class 1 integrons from human commensal bacteria in a hospital intensive care unit. Antimicrobial agents and chemotherapy. 2011. 55. pp. 3939-43. doi: 10.1128/AAC.01831-10. PMID: 21628540.
- ↑ Arakawa et al.. A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrobial agents and chemotherapy. 1995. 39. pp. 1612-5. doi: 10.1128/AAC.39.7.1612. PMID: 7492116.
- ↑ Collis et al.. Characterization of the class 3 integron and the site-specific recombination system it determines. Journal of bacteriology. 2002. 184. pp. 3017-26. doi: 10.1128/JB.184.11.3017-3026.2002. PMID: 12003943.
- ↑ Oosterkamp et al.. Genome analysis and physiological comparison of Alicycliphilus denitrificans strains BC and K601(T.). PloS one. 2013. 8. pp. e66971. doi: 10.1371/journal.pone.0066971. PMID: 23825601.
- ↑ 78.0 78.1 Nicolas et al.. The Tn3-family of Replicative Transposons. Microbiology spectrum. 2015. 3. doi: 10.1128/microbiolspec.MDNA3-0060-2014. PMID: 26350313.
- ↑ Barkay et al.. A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environmental microbiology. 2010. 12. pp. 2904-17. doi: 10.1111/j.1462-2920.2010.02260.x. PMID: 20545753.
- ↑ Boyd & Barkay. The mercury resistance operon: from an origin in a geothermal environment to an efficient detoxification machine. Frontiers in microbiology. 2012. 3. pp. 349. doi: 10.3389/fmicb.2012.00349. PMID: 23087676.
- ↑ HAMLET N, LANDALE E, DAVIS B, SUMMERS A. Roles of the Tn2l merT, merP, and merC Gene Products in Mercury Resistance and Mercury Binding. J Bacteriol. 1992;174:6377–6385.
- ↑ 82.0 82.1 Stanisich V, Arwas R, Bennett PM, De La Cruz F. Characterization of Pseudomonas Mercury - resistance Transposon Tn502, Which Has a Preferred Insertion Site in RP1. J Gen Microbiol. 1989;135:2909–2915.
- ↑ Roberts et al.. Genetic characterization of the stabilizing functions of a region of broad-host-range plasmid RK2. Journal of bacteriology. 1990. 172. pp. 6204-16. doi: 10.1128/jb.172.11.6204-6216.1990. PMID: 2121707.
- ↑ Gerlitz et al.. Partitioning of broad-host-range plasmid RP4 is a complex system involving site-specific recombination. Journal of bacteriology. 1990. 172. pp. 6194-203. doi: 10.1128/jb.172.11.6194-6203.1990. PMID: 2172207.
- ↑ Eberl et al.. Analysis of the multimer resolution system encoded by the parCBA operon of broad-host-range plasmid RP4. Molecular microbiology. 1994. 12. pp. 131-41. doi: 10.1111/j.1365-2958.1994.tb01002.x. PMID: 8057833.
- ↑ 86.0 86.1 86.2 86.3 86.4 86.5 Kamali-Moghaddam & Sundström. Transposon targeting determined by resolvase. FEMS microbiology letters. 2000. 186. pp. 55-9. doi: 10.1111/j.1574-6968.2000.tb09081.x. PMID: 10779712.
- ↑ 87.0 87.1 Sinclair & Holloway. A chromosomally located transposon in Pseudomonas aeruginosa. Journal of bacteriology. 1982. 151. pp. 569-79. doi: 10.1128/jb.151.2.569-579.1982. PMID: 6284702.
- ↑ Partridge et al.. Family of class 1 integrons related to In4 from Tn1696. Antimicrobial agents and chemotherapy. 2001. 45. pp. 3014-20. doi: 10.1128/AAC.45.11.3014-3020.2001. PMID: 11600350.
- ↑ Brown & Willetts. A physical and genetic map of the IncN plasmid R46. Plasmid. 1981. 5. pp. 188-201. doi: 10.1016/0147-619x(81)90020-2. PMID: 6264522.
- ↑ Walker. Plasmid (pKM101)-mediated enhancement of repair and mutagenesis: dependence on chromosomal genes in Escherichia coli K-12. Molecular & general genetics : MGG. 1977. 152. pp. 93-103. doi: 10.1007/BF00264945. PMID: 325391.
- ↑ Tosini et al.. The uvp1 gene on the R46 plasmid encodes a resolvase that catalyzes site-specific resolution involving the 5'-conserved segment of the adjacent integron In1. Molecular & general genetics : MGG. 1998. 258. pp. 404-11. doi: 10.1007/s004380050748. PMID: 9648746.
- ↑ Dodd & Bennett. R46 encodes a site-specific recombination system interchangeable with the resolution function of TnA. Plasmid. 1983. 9. pp. 247-61. doi: 10.1016/0147-619x(83)90003-3. PMID: 6306704.
- ↑ 93.0 93.1 Dodd & Bennett. Location of the site-specific recombination system of R46: a function necessary for plasmid maintenance. Journal of general microbiology. 1986. 132. pp. 1009-20. doi: 10.1099/00221287-132-4-1009. PMID: 3020154.
- ↑ Dodd HM, Bennett PM. The R46 Site-specific Recombination System is a Homoiogue of the Tn3 and gamma delta (Tn1000) Cointegrate Resolution System.
- ↑ 95.0 95.1 95.2 95.3 95.4 95.5 Minakhina et al.. Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Molecular microbiology. 1999. 33. pp. 1059-68. doi: 10.1046/j.1365-2958.1999.01548.x. PMID: 10476039.
- ↑ Kholodii et al.. Tn5044, a novel Tn3 family transposon coding for temperature-sensitive mercury resistance. Research in microbiology. 2000. 151. pp. 291-302. doi: 10.1016/s0923-2508(00)00149-2. PMID: 10875286.
- ↑ Allmeier et al.. Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene. 1992. 111. pp. 11-20. doi: 10.1016/0378-1119(92)90597-i. PMID: 1312499.
- ↑ Diver et al.. DNA sequences of and complementation by the tnpR genes of Tn21, Tn501 and Tn1721. Molecular & general genetics : MGG. 1983. 191. pp. 189-93. doi: 10.1007/BF00334812. PMID: 6312271.
- ↑ 99.0 99.1 Peters. Tn7. Microbiology spectrum. 2014. 2. doi: 10.1128/microbiolspec.MDNA3-0010-2014. PMID: 26104363.
- ↑ Peters & Craig. Tn7 recognizes transposition target structures associated with DNA replication using the DNA-binding protein TnsE. Genes & development. 2001. 15. pp. 737-47. doi: 10.1101/gad.870201. PMID: 11274058.
- ↑ Kuduvalli et al.. Target DNA structure plays a critical role in Tn7 transposition. The EMBO journal. 2001. 20. pp. 924-32. doi: 10.1093/emboj/20.4.924. PMID: 11179236.
- ↑ Hickman et al.. Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Molecular cell. 2000. 5. pp. 1025-34. doi: 10.1016/s1097-2765(00)80267-1. PMID: 10911996.
- ↑ Sarnovsky et al.. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. The EMBO journal. 1996. 15. pp. 6348-61. PMID: 8947057.
- ↑ Bainton et al.. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991. 65. pp. 805-16. doi: 10.1016/0092-8674(91)90388-f. PMID: 1645619.
- ↑ May & Craig. Switching from cut-and-paste to replicative Tn7 transposition. Science (New York, N.Y.). 1996. 272. pp. 401-4. doi: 10.1126/science.272.5260.401. PMID: 8602527.
How to Cite?
TnPedia Team. (2025). TnPedia – Chapter: The Tn402 Family of Prokaryotic Transposons. Zenodo. https://doi.org/10.5281/zenodo.15566551