General Information/Non-autonomous IS derivatives: Difference between revisions
m Text replacement - "</pubmed>" to "" |
No edit summary |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== Non-autonomous Elements == | |||
Many prokaryotic genomes are littered with IS fragments and small, non-autonomous IS derivatives whose transposition can, in principle, be catalyzed in trans by the Tpase of a related complete IS [[:Image:1.13.1.png|(Fig.8.1)]]. Structures called MITEs ('''M'''iniature '''I'''nverted repeat '''T'''ransposable '''E'''lements), first identified in ''Neisseria''<ref name=":0">{{#pmid:2842323}} | |||
</ref> are found in many organisms including plants<ref>{{#pmid:12618411}}</ref><ref name=":1">{{#pmid:11756687}}</ref><ref>{{#pmid:1332797}}</ref><ref>{{#pmid:8061524}}</ref>, insects<ref>{{#pmid:16507338}}</ref><ref>{{#pmid:16919158}}</ref>, fungi<ref>{{#pmid:17179071}}</ref> bacteria <ref name=":0" /><ref>{{#pmid:19393167}}</ref><ref>{{#pmid:22016774}}</ref><ref>{{#pmid:31085513}}</ref><ref>{{#pmid:26442174}}</ref> and archaea<ref>{{#pmid:15046567}}</ref><ref name=":2">{{#pmid:11814653}}</ref><ref>{{#pmid:16935554}}</ref> are related to IS with DDE Tpases<ref name=":2" /><ref name=":3">{{#pmid:17347521}} | |||
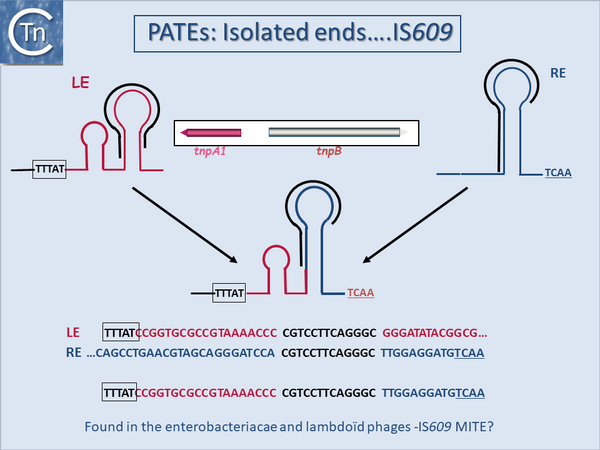
</ref>. Among those derived from IS with DDE transposases, representatives of the [[IS Families/IS1 family|IS''1'']], [[IS Families/IS4 and related families|IS''4'']]'','' [[IS Families/IS5 and related IS1182 families|IS''5'']], [[IS Families/IS6 family|IS''6'']], and even [[Transposons families/Tn3 family|Tn''3'' family]] members such as [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISRf1 IS''Rf1''] from ''[[wikipedia:Sinorhizobium_fredii|Sinorhizobium fredii]]'' have been identified. [[Image:1.18.1.png|thumb|600x600px|'''Fig.13.1.''' PATEs ('''P'''alindrome-'''A'''ssociated '''T'''ransposable '''E'''lements) isolated ends from [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS609 IS''609'']. | |||
The particular case of MITES derived from [[IS Families/IS200-IS605 family|IS''200/IS605'' family]] members. Top: The IS ends with their essential secondary structures are shown in red (left end) and blue (right end). The boxed (left) sequence represents the conserved target sequence and underlined (right) sequence at the right end are sequences recognized by guide sequences at the foot of the secondary structures necessary for directing cleavage. The inset represents a full-sized IS with its transposase ''tnpA1'' (red) and the accessory gene ''tnpB'' (pale green). The actual DNA sequences are shown at the bottom of the figure.|alt=|center]]They are generally less than 300 bp long, include appropriately oriented left and right terminal IR but no Tpase and generally generate flanking DR. MITEs are probably derived from IS by internal deletion. Some carry short non-coding sequences between these IRs which may or may not be IS-derived. MITEs are considered to be non-autonomous transposable elements mobilizable in trans by Tpases of full-length parental genomic copies. They were first identified in plants<ref name=":1" /><ref>{{#pmid:15020481}}</ref><ref>{{#pmid:15831788}}</ref><ref>{{#pmid:17578919}}</ref> and are related to Tc/mariner elements (distantly related to bacterial [[IS Families/IS630 family|IS''630'' family]]). [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS630 IS''630'']-related MITEs were also the first described bacterial examples<ref name=":0" /><ref>{{#pmid:11707339}}</ref><ref>{{#pmid:12095618}}</ref><ref>{{#pmid:21283790}}</ref><ref>{{#pmid:10537186}}</ref>. MITEs showing similarities to other IS and transposon families are observed in bacteria and archaea. Those related to the [[Transposons families/Tn3 family|Tn''3'' family]] have been called TIMEs<ref>{{#pmid:25121765}}</ref>. | |||
MITE-like structures related to elements with other types of transposase have also been identified. Among these are [[IS Families/IS200-IS605 family|IS''200''/IS''605'' family]] derivatives<ref name=":3" />; ([https://scholar.google.fr/citations?user=WHAtfqcAAAAJ&hl=fr P. Siguier], unpublished) [[:Image:1.18.1.png|(Fig.13.1)]] now called PATEs ('''P'''alindrome-'''A'''ssociated '''T'''ransposable '''E'''lements)<ref>{{#pmid:21701686}}</ref> reflecting the sub-terminal secondary structures which constitute the ends of these IS. Although originally observed in the Archaea as derivatives of known [[IS Families/IS200-IS605 family|IS''200''/IS''605'' family]] members, they have also been observed in certain cyanobacteria and ''[[wikipedia:Salmonella|Salmonella]]'' ([https://scholar.google.fr/citations?user=WHAtfqcAAAAJ&hl=fr P. Siguier] unpublished). They presumably represent decay products that appear quite frequently for this IS family. | MITE-like structures related to elements with other types of transposase have also been identified. Among these are [[IS Families/IS200-IS605 family|IS''200''/IS''605'' family]] derivatives<ref name=":3" />; ([https://scholar.google.fr/citations?user=WHAtfqcAAAAJ&hl=fr P. Siguier], unpublished) [[:Image:1.18.1.png|(Fig.13.1)]] now called PATEs ('''P'''alindrome-'''A'''ssociated '''T'''ransposable '''E'''lements)<ref>{{#pmid:21701686}}</ref> reflecting the sub-terminal secondary structures which constitute the ends of these IS. Although originally observed in the Archaea as derivatives of known [[IS Families/IS200-IS605 family|IS''200''/IS''605'' family]] members, they have also been observed in certain cyanobacteria and ''[[wikipedia:Salmonella|Salmonella]]'' ([https://scholar.google.fr/citations?user=WHAtfqcAAAAJ&hl=fr P. Siguier] unpublished). They presumably represent decay products that appear quite frequently for this IS family. | ||
Full-length copies of the parental IS may not be available or may be so divergent as to escape detection in standard [https://blast.ncbi.nlm.nih.gov/Blast.cgi BLAST analysi]s. This is the case for certain MITEs from the archaea <ref>{{#pmid: | Full-length copies of the parental IS may not be available or may be so divergent as to escape detection in standard [https://blast.ncbi.nlm.nih.gov/Blast.cgi BLAST analysi]s. This is the case for certain MITEs from the archaea <ref>{{#pmid:15612937}}</ref> and is probably also true for the bacteria. Their detection and analysis are therefore arduous. | ||
Another group of IS derivatives related to MITEs are called MICs ('''M'''obile '''I'''nsertion '''C'''assette)<ref name=":4">{{#pmid:15228527}} | |||
</ref><ref>{{#pmid:10320586}}</ref> [[:Image:1.13.1.png|(Fig.8.1)]]. These, like MITEs, are flanked by IR, do not include a Tpase gene and generate flanking DR. They carry various coding sequences, and are present in relatively low copy number. The [[IS Families/IS4 and related families#IS231|IS''231'' sub-group]] of the large [[IS Families/IS4 and related families|IS''4'' family]] includes examples of many of these IS-derivatives (canonical ISs, MITEs, MICs and tIS)<ref name=":4" /><ref>{{#pmid:18215304}}</ref>. Tn''3''-derived MICs appear to be particularly prevalent in ''Xanthomonas'' where they include TALE ('''T'''ranscriptional '''A'''ctivator '''L'''ike '''E'''lements) genes involved in ''Xanthomonas'' pathogenicity in plants <ref>{{#pmid:25691597}}</ref> | </ref><ref>{{#pmid:10320586}}</ref> [[:Image:1.13.1.png|(Fig.8.1)]]. These, like MITEs, are flanked by IR, do not include a Tpase gene and generate flanking DR. They carry various coding sequences, and are present in relatively low copy number. The [[IS Families/IS4 and related families#IS231|IS''231'' sub-group]] of the large [[IS Families/IS4 and related families|IS''4'' family]] includes examples of many of these IS-derivatives (canonical ISs, MITEs, MICs and tIS)<ref name=":4" /><ref>{{#pmid:18215304}}</ref>. Tn''3''-derived MICs appear to be particularly prevalent in ''Xanthomonas'' where they include TALE ('''T'''ranscriptional '''A'''ctivator '''L'''ike '''E'''lements) genes involved in ''Xanthomonas'' pathogenicity in plants <ref>{{#pmid:25691597}}</ref> | ||
==Bibliography== | |||
{{Reflist|32em}} | |||
== How to Cite? == | |||
TnPedia Team. (2025). TnPedia: General Information on Prokaryotic Elements. Zenodo. https://doi.org/10.5281/zenodo.15548171 | |||
[[File:General_Info-badge.png|link=https://doi.org/10.5281/zenodo.15548171|DOI badge]]<hr> | |||
<hr> | |||
{{TnPedia}} | {{TnPedia}} | ||
Latest revision as of 11:35, 1 June 2025
Non-autonomous Elements
Many prokaryotic genomes are littered with IS fragments and small, non-autonomous IS derivatives whose transposition can, in principle, be catalyzed in trans by the Tpase of a related complete IS (Fig.8.1). Structures called MITEs (Miniature Inverted repeat Transposable Elements), first identified in Neisseria[1] are found in many organisms including plants[2][3][4][5], insects[6][7], fungi[8] bacteria [1][9][10][11][12] and archaea[13][14][15] are related to IS with DDE Tpases[14][16]. Among those derived from IS with DDE transposases, representatives of the IS1, IS4, IS5, IS6, and even Tn3 family members such as ISRf1 from Sinorhizobium fredii have been identified.
They are generally less than 300 bp long, include appropriately oriented left and right terminal IR but no Tpase and generally generate flanking DR. MITEs are probably derived from IS by internal deletion. Some carry short non-coding sequences between these IRs which may or may not be IS-derived. MITEs are considered to be non-autonomous transposable elements mobilizable in trans by Tpases of full-length parental genomic copies. They were first identified in plants[3][17][18][19] and are related to Tc/mariner elements (distantly related to bacterial IS630 family). IS630-related MITEs were also the first described bacterial examples[1][20][21][22][23]. MITEs showing similarities to other IS and transposon families are observed in bacteria and archaea. Those related to the Tn3 family have been called TIMEs[24].
MITE-like structures related to elements with other types of transposase have also been identified. Among these are IS200/IS605 family derivatives[16]; (P. Siguier, unpublished) (Fig.13.1) now called PATEs (Palindrome-Associated Transposable Elements)[25] reflecting the sub-terminal secondary structures which constitute the ends of these IS. Although originally observed in the Archaea as derivatives of known IS200/IS605 family members, they have also been observed in certain cyanobacteria and Salmonella (P. Siguier unpublished). They presumably represent decay products that appear quite frequently for this IS family.
Full-length copies of the parental IS may not be available or may be so divergent as to escape detection in standard BLAST analysis. This is the case for certain MITEs from the archaea [26] and is probably also true for the bacteria. Their detection and analysis are therefore arduous.
Another group of IS derivatives related to MITEs are called MICs (Mobile Insertion Cassette)[27][28] (Fig.8.1). These, like MITEs, are flanked by IR, do not include a Tpase gene and generate flanking DR. They carry various coding sequences, and are present in relatively low copy number. The IS231 sub-group of the large IS4 family includes examples of many of these IS-derivatives (canonical ISs, MITEs, MICs and tIS)[27][29]. Tn3-derived MICs appear to be particularly prevalent in Xanthomonas where they include TALE (Transcriptional Activator Like Elements) genes involved in Xanthomonas pathogenicity in plants [30]
Bibliography
- ↑ 1.0 1.1 1.2 Correia et al.. A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. The Journal of biological chemistry. 1988. 263. pp. 12194-8. PMID: 2842323.
- ↑ Feschotte et al.. Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with stowaway miniature inverted repeat transposable elements (MITEs). Genetics. 2003. 163. pp. 747-58. doi: 10.1093/genetics/163.2.747. PMID: 12618411.
- ↑ 3.0 3.1 Feschotte & Wessler. Mariner-like transposases are widespread and diverse in flowering plants. Proceedings of the National Academy of Sciences of the United States of America. 2002. 99. pp. 280-5. doi: 10.1073/pnas.022626699. PMID: 11756687.
- ↑ Bureau & Wessler. Tourist: a large family of small inverted repeat elements frequently associated with maize genes. The Plant cell. 1992. 4. pp. 1283-94. doi: 10.1105/tpc.4.10.1283. PMID: 1332797.
- ↑ Bureau & Wessler. Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. The Plant cell. 1994. 6. pp. 907-16. doi: 10.1105/tpc.6.6.907. PMID: 8061524.
- ↑ Palomeque et al.. Detection of a mariner-like element and a miniature inverted-repeat transposable element (MITE) associated with the heterochromatin from ants of the genus Messor and their possible involvement for satellite DNA evolution. Gene. 2006. 371. pp. 194-205. doi: 10.1016/j.gene.2005.11.032. PMID: 16507338.
- ↑ Quesneville et al.. P elements and MITE relatives in the whole genome sequence of Anopheles gambiae. BMC genomics. 2006. 7. pp. 214. doi: 10.1186/1471-2164-7-214. PMID: 16919158.
- ↑ Dufresne et al.. Transposition of a fungal miniature inverted-repeat transposable element through the action of a Tc1-like transposase. Genetics. 2007. 175. pp. 441-52. doi: 10.1534/genetics.106.064360. PMID: 17179071.
- ↑ Chen et al.. MUST: a system for identification of miniature inverted-repeat transposable elements and applications to Anabaena variabilis and Haloquadratum walsbyi. Gene. 2009. 436. pp. 1-7. doi: 10.1016/j.gene.2009.01.019. PMID: 19393167.
- ↑ Bardaji et al.. Miniature transposable sequences are frequently mobilized in the bacterial plant pathogen Pseudomonas syringae pv. phaseolicola. PloS one. 2011. 6. pp. e25773. doi: 10.1371/journal.pone.0025773. PMID: 22016774.
- ↑ Kieffer et al.. Functional Characterization of a Miniature Inverted Transposable Element at the Origin of mcr-5 Gene Acquisition in Escherichia coli. Antimicrobial agents and chemotherapy. 2019. 63. doi: 10.1128/AAC.00559-19. PMID: 31085513.
- ↑ Szuplewska et al.. Autonomous and non-autonomous Tn3-family transposons and their role in the evolution of mobile genetic elements. Mobile genetic elements. 2014. 4. pp. 1-4. doi: 10.1080/2159256X.2014.998537. PMID: 26442174.
- ↑ Brügger et al.. Shuffling of Sulfolobus genomes by autonomous and non-autonomous mobile elements. Biochemical Society transactions. 2004. 32. pp. 179-83. doi: 10.1042/bst0320179. PMID: 15046567.
- ↑ 14.0 14.1 Brügger et al.. Mobile elements in archaeal genomes. FEMS microbiology letters. 2002. 206. pp. 131-41. doi: 10.1111/j.1574-6968.2002.tb10999.x. PMID: 11814653.
- ↑ Siguier et al.. Insertion sequences in prokaryotic genomes. Current opinion in microbiology. 2006. 9. pp. 526-31. doi: 10.1016/j.mib.2006.08.005. PMID: 16935554.
- ↑ 16.0 16.1 Filée et al.. Insertion sequence diversity in archaea. Microbiology and molecular biology reviews : MMBR. 2007. 71. pp. 121-57. doi: 10.1128/MMBR.00031-06. PMID: 17347521.
- ↑ Zhang et al.. PIF- and Pong-like transposable elements: distribution, evolution and relationship with Tourist-like miniature inverted-repeat transposable elements. Genetics. 2004. 166. pp. 971-86. doi: 10.1534/genetics.166.2.971. PMID: 15020481.
- ↑ Feschotte et al.. DNA-binding specificity of rice mariner-like transposases and interactions with Stowaway MITEs. Nucleic acids research. 2005. 33. pp. 2153-65. doi: 10.1093/nar/gki509. PMID: 15831788.
- ↑ Yang et al.. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2007. 104. pp. 10962-7. doi: 10.1073/pnas.0702080104. PMID: 17578919.
- ↑ Mazzone et al.. Whole-genome organization and functional properties of miniature DNA insertion sequences conserved in pathogenic Neisseriae. Gene. 2001. 278. pp. 211-22. doi: 10.1016/s0378-1119(01)00725-9. PMID: 11707339.
- ↑ Buisine et al.. Transposon-like Correia elements: structure, distribution and genetic exchange between pathogenic Neisseria sp. FEBS letters. 2002. 522. pp. 52-8. doi: 10.1016/s0014-5793(02)02882-x. PMID: 12095618.
- ↑ Siddique et al.. The transposon-like Correia elements encode numerous strong promoters and provide a potential new mechanism for phase variation in the meningococcus. PLoS genetics. 2011. 7. pp. e1001277. doi: 10.1371/journal.pgen.1001277. PMID: 21283790.
- ↑ Oggioni & Claverys. Repeated extragenic sequences in prokaryotic genomes: a proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology (Reading, England). 1999. 145 ( Pt 10). pp. 2647-53. doi: 10.1099/00221287-145-10-2647. PMID: 10537186.
- ↑ Szuplewska et al.. Mobility and generation of mosaic non-autonomous transposons by Tn3-derived inverted-repeat miniature elements (TIMEs). PloS one. 2014. 9. pp. e105010. doi: 10.1371/journal.pone.0105010. PMID: 25121765.
- ↑ Dyall-Smith et al.. Haloquadratum walsbyi: limited diversity in a global pond. PloS one. 2011. 6. pp. e20968. doi: 10.1371/journal.pone.0020968. PMID: 21701686.
- ↑ Blount & Grogan. New insertion sequences of Sulfolobus: functional properties and implications for genome evolution in hyperthermophilic archaea. Molecular microbiology. 2005. 55. pp. 312-25. doi: 10.1111/j.1365-2958.2004.04391.x. PMID: 15612937.
- ↑ 27.0 27.1 De Palmenaer et al.. IS231-MIC231 elements from Bacillus cereus sensu lato are modular. Molecular microbiology. 2004. 53. pp. 457-67. doi: 10.1111/j.1365-2958.2004.04146.x. PMID: 15228527.
- ↑ Chen et al.. MIC231, a naturally occurring mobile insertion cassette from Bacillus cereus. Molecular microbiology. 1999. 32. pp. 657-68. doi: 10.1046/j.1365-2958.1999.01388.x. PMID: 10320586.
- ↑ De Palmenaer et al.. IS4 family goes genomic. BMC evolutionary biology. 2008. 8. pp. 18. doi: 10.1186/1471-2148-8-18. PMID: 18215304.
- ↑ Ferreira et al.. A TALE of transposition: Tn3-like transposons play a major role in the spread of pathogenicity determinants of Xanthomonas citri and other xanthomonads. mBio. 2015. 6. pp. e02505-14. doi: 10.1128/mBio.02505-14. PMID: 25691597.
How to Cite?
TnPedia Team. (2025). TnPedia: General Information on Prokaryotic Elements. Zenodo. https://doi.org/10.5281/zenodo.15548171