IS Families/ISAzo13 family: Difference between revisions
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
{{Reflist|32em}} | {{Reflist|32em}} | ||
<br /> | <br /> | ||
== How to Cite? == | |||
Zenodo. TnCentral and TnPedia Team. (2025). TnPedia: IS''XXXXXX'' Family of Prokaryotic Insertion Sequences. Zenodo. https://doi.org/XXXXXX/zenodo.YYYYYYYY | |||
[[File:IS3-zenodo.YYYYYYYY.png|link=https://doi.org/XXXXXX/zenodo.YYYYYYYY|DOI badge]] | |||
<hr> | <hr> | ||
{{TnPedia}} | {{TnPedia}} | ||
Revision as of 16:24, 10 June 2025
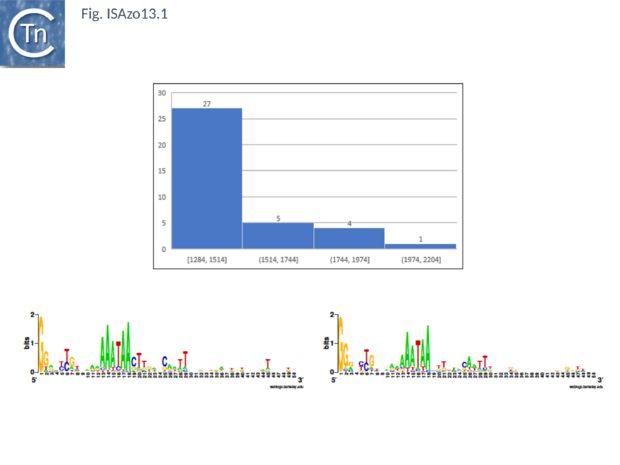
This family, represented by 37 members in ISfinder emerged from the ISNCY orphan group. It is based on both Tpase and IR sequence similarities (Fig.ISAzo13). Insertion generates a 3bp AT-rich DR and the ends have a consensus GGa/g. Their Tpases are highly conserved with a probable DDE motif and an HTH motif at the N-terminus which could function as a DNA binding domain. Two members encode two orfs with a possible PRF (Programmed Transcriptional Frameshifting) motif of 8 or 9 A while the other members encode a unique orf which includes a triple lysine at the equivalent position (Gourbeyre, unpublished)[1].
Bibliography
- ↑ Siguier et al.. ISfinder: the reference centre for bacterial insertion sequences. Nucleic acids research. 2006. 34. pp. D32-6. doi: 10.1093/nar/gkj014. PMID: 16381877.
How to Cite?
Zenodo. TnCentral and TnPedia Team. (2025). TnPedia: ISXXXXXX Family of Prokaryotic Insertion Sequences. Zenodo. https://doi.org/XXXXXX/zenodo.YYYYYYYY