General Information/IS and Gene Expression: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
'''<big>A</big>'''nother important aspect of IS impact on their bacterial hosts is their ability to modulate gene expression. In addition to acting as vectors for gene transmission from one replicon to another in the form composite transposons (two IS flanking any gene; [[:Image:1.2.3.png|Fig.2.3]]) and tIS [[:Image:1.13.1.png|(Fig.8.1)]] and their ability to interrupt genes, it has been known for some time<ref><nowiki><pubmed>1101028</pubmed></nowiki></ref><ref name=":0"><pubmed>6271458</pubmed></nowiki></ref> that IS can also activate gene expression. This capacity has recently received much attention due to the increase in resistance to various antibacterials<ref name=":1"><pubmed>16952941</pubmed></nowiki></ref><ref name=":2"><pubmed>12923109</pubmed></nowiki></ref><ref name=":3"><pubmed>23158541</pubmed></nowiki></ref>, a worrying public health threat<ref><nowiki><pubmed>23887414</pubmed></nowiki></ref><ref><nowiki><pubmed>23887415</pubmed></nowiki></ref>. | '''<big>A</big>'''nother important aspect of IS impact on their bacterial hosts is their ability to modulate gene expression. In addition to acting as vectors for gene transmission from one replicon to another in the form composite transposons (two IS flanking any gene; [[:Image:1.2.3.png|Fig.2.3]]) and tIS [[:Image:1.13.1.png|(Fig.8.1)]] and their ability to interrupt genes, it has been known for some time<ref><nowiki><pubmed>1101028</pubmed></nowiki></ref><ref name=":0"><pubmed>6271458</pubmed></nowiki></ref> that IS can also activate gene expression. This capacity has recently received much attention due to the increase in resistance to various antibacterials<ref name=":1"><pubmed>16952941</pubmed></nowiki></ref><ref name=":2"><pubmed>12923109</pubmed></nowiki></ref><ref name=":3"><pubmed>23158541</pubmed></nowiki></ref>, a worrying public health threat<ref><nowiki><pubmed>23887414</pubmed></nowiki></ref><ref><nowiki><pubmed>23887415</pubmed></nowiki></ref>. | ||
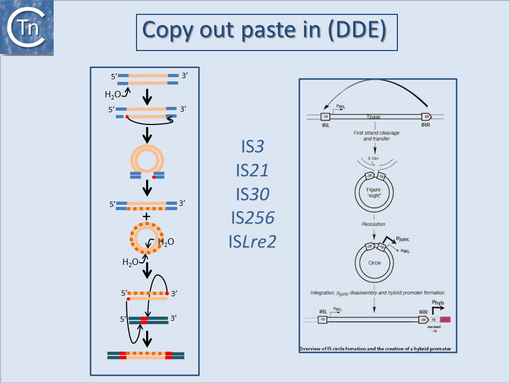
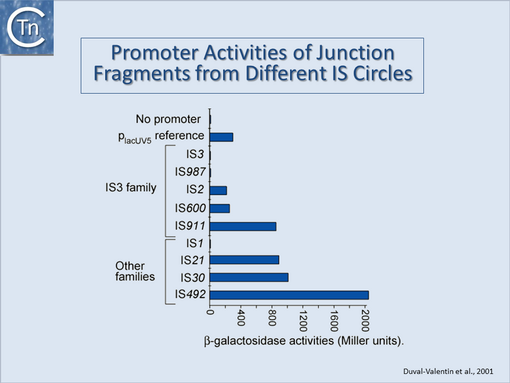
They can accomplish this in two ways: either by providing internal promoters whose transcripts escape into neighboring DNA<ref name=":0" /><ref name=":4"><pubmed>6292860</pubmed></nowiki></ref><ref><nowiki><pubmed>6260746</pubmed></nowiki></ref><ref><nowiki><pubmed>6311437</pubmed></nowiki></ref> or by hybrid promoter formation. Many IS carry -35 promoter components oriented towards the flanking DNA [[:Image:1.24.1.png|(Fig.18.1)]]. In a number of cases this plays an important part in their transposition since a significant number of IS | They can accomplish this in two ways: either by providing internal promoters whose transcripts escape into neighboring DNA<ref name=":0" /><ref name=":4"><pubmed>6292860</pubmed></nowiki></ref><ref><nowiki><pubmed>6260746</pubmed></nowiki></ref><ref><nowiki><pubmed>6311437</pubmed></nowiki></ref> or by hybrid promoter formation. Many IS carry -35 promoter components oriented towards the flanking DNA [[:Image:1.24.1.png|(Fig.18.1)]]. In a number of cases this plays an important part in their transposition since a significant number of IS transposes using an excised transposon circle [[:Image:1.24.1.png|(Fig.18.1)]] with abutted left and right ends. For these IS, the other end carries a -10 element oriented inwards towards the Tpase gene. Together with the -35, this generates a strong promoter on formation of the circle junction to drive Tpase expression required for catalysis of integration [[:Image:1.24.2.png|(Fig.18.2)]] <ref><nowiki><pubmed>26350305</pubmed></nowiki></ref><ref><nowiki><pubmed>9214651</pubmed></nowiki></ref><ref><nowiki><pubmed>10438765</pubmed></nowiki></ref><ref><nowiki><pubmed>11598022</pubmed></nowiki></ref>. Thus if integration occurs next to a resident -10 sequence, the IS -35 sequence can contribute to a hybrid promoter to drive expression of neighboring genes [see <ref name=":5"><pubmed>3029382</pubmed></nowiki></ref>]. At present this phenomenon had been reported to occur with over 30 different IS in more than 17 bacterial species<ref><nowiki><pubmed>17223624</pubmed></nowiki></ref><ref><nowiki><pubmed>24499397</pubmed></nowiki></ref> ([[General Information/IS and Gene Expression#IS and Gene Expression|Table IS and Gene Expression below]]). Indeed, specific vector plasmids have been designed to identify activating insertions (e.g. <ref><nowiki><pubmed>8863735</pubmed></nowiki></ref>). | ||
IS activity can affect efflux mechanisms resulting in increased resistance: IS''1'' or IS''10'' insertion can up-regulate the [[wikipedia:Efflux_(microbiology)|AcrAB-TolC]] pump in ''[[wikipedia:Salmonella_enterica|Salmonella enterica]]''<ref name=":6"><pubmed>15616308</pubmed></nowiki></ref>; IS''1'' or IS''2'' insertion upstream of AcrEF<ref name=":7"><pubmed>11302812</pubmed></nowiki></ref><ref><nowiki><pubmed>11274125</pubmed></nowiki></ref> and IS''186'' insertional inactivation of the AcrAB repressor, AcrR, in ''[[wikipedia:Escherichia_coli|Escherichia coli]] <ref name=":7" />'', all lead to increased resistance to [[wikipedia:Quinolone_antibiotic|fluoroquinolones]]. Insertional inactivation of specific [[wikipedia:Porin_(protein)|porins]] can also play a significant role<ref><nowiki><pubmed>15212803</pubmed></nowiki></ref>.<center> | IS activity can affect efflux mechanisms resulting in increased resistance: IS''1'' or IS''10'' insertion can up-regulate the [[wikipedia:Efflux_(microbiology)|AcrAB-TolC]] pump in ''[[wikipedia:Salmonella_enterica|Salmonella enterica]]''<ref name=":6"><pubmed>15616308</pubmed></nowiki></ref>; IS''1'' or IS''2'' insertion upstream of AcrEF<ref name=":7"><pubmed>11302812</pubmed></nowiki></ref><ref><nowiki><pubmed>11274125</pubmed></nowiki></ref> and IS''186'' insertional inactivation of the AcrAB repressor, AcrR, in ''[[wikipedia:Escherichia_coli|Escherichia coli]] <ref name=":7" />'', all lead to increased resistance to [[wikipedia:Quinolone_antibiotic|fluoroquinolones]]. Insertional inactivation of specific [[wikipedia:Porin_(protein)|porins]] can also play a significant role<ref><nowiki><pubmed>15212803</pubmed></nowiki></ref>.<center> | ||
| Line 19: | Line 19: | ||
! scope="col" |IS family||IS name||Mechanism||Gene(s) affected||Organism||Reference||Clinical/Experimental | ! scope="col" |IS family||IS name||Mechanism||Gene(s) affected||Organism||Reference||Clinical/Experimental | ||
|- | |- | ||
| rowspan="3" |[[IS Families/IS1 family|IS<i>1</i>]]|| rowspan="3" |IS<i>1</i>||Cointegrate||<i>Em<sup>R</sup></i>|| rowspan="2" |<i>Escherichia coli</i>||<ref name=":8"><pubmed>6283551</pubmed></nowiki></ref>||C? | | rowspan="3" |[[IS Families/IS1 family|IS<i>1</i>]]|| rowspan="3" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1A IS<i>1</i>]||Cointegrate||<i>Em<sup>R</sup></i>|| rowspan="2" |<i>Escherichia coli</i>||<ref name=":8"><pubmed>6283551</pubmed></nowiki></ref>||C? | ||
|- | |- | ||
|Copy-paste circles||<i>bla<sub>TEM-1</sub></i>||<ref name=":5" />||E | |Copy-paste circles||<i>bla<sub>TEM-1</sub></i>||<ref name=":5" />||E | ||
| Line 25: | Line 25: | ||
|—||<i>acrEF</i> pump||<i>Salmonella enterica</i>||<ref name=":6" />||E | |—||<i>acrEF</i> pump||<i>Salmonella enterica</i>||<ref name=":6" />||E | ||
|- | |- | ||
| rowspan="12" |[[IS Families/IS3 family|IS<i>3</i>]]|| rowspan="6" |IS<i>2</i>|| rowspan="6" |Copy-paste circles||<i>gal</i>|| rowspan="6" |<i>Escherichia coli</i>||<ref><nowiki><pubmed>4610339</pubmed></nowiki></ref><ref><nowiki><pubmed>339095</pubmed></nowiki></ref>||E | | rowspan="12" |[[IS Families/IS3 family|IS<i>3</i>]]|| rowspan="6" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS2 IS<i>2</i>]|| rowspan="6" |Copy-paste circles||<i>gal</i>|| rowspan="6" |<i>Escherichia coli</i>||<ref><nowiki><pubmed>4610339</pubmed></nowiki></ref><ref><nowiki><pubmed>339095</pubmed></nowiki></ref>||E | ||
|- | |- | ||
|<i>gal</i>|||<ref name=":8" /><ref name=":0" />||E | |<i>gal</i>|||<ref name=":8" /><ref name=":0" />||E | ||
| Line 37: | Line 37: | ||
|<i>acrEF</i> pump||E | |<i>acrEF</i> pump||E | ||
|- | |- | ||
| rowspan="3" |IS<i>3</i>|| rowspan="3" |Copy-paste circles||<i>argE</i>|| rowspan="3" |<i>Escherichia coli</i>||<ref name=":0" />||E | | rowspan="3" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS3 IS<i>3</i>]|| rowspan="3" |Copy-paste circles||<i>argE</i>|| rowspan="3" |<i>Escherichia coli</i>||<ref name=":0" />||E | ||
|- | |- | ||
|<i>argE</i>||<ref name=":4" />||E | |<i>argE</i>||<ref name=":4" />||E | ||
| Line 43: | Line 43: | ||
|<i>citT</i>||<ref><nowiki><pubmed>22992527</pubmed></nowiki></ref>||E | |<i>citT</i>||<ref><nowiki><pubmed>22992527</pubmed></nowiki></ref>||E | ||
|- | |- | ||
|IS<i>981</i>||Copy-paste circles||<i>ldhB</i>||<i>Lactococcus lactis</i>||<ref><nowiki><pubmed>12867459</pubmed></nowiki></ref>||E | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS981 IS<i>981</i>]||Copy-paste circles||<i>ldhB</i>||<i>Lactococcus lactis</i>||<ref><nowiki><pubmed>12867459</pubmed></nowiki></ref>||E | ||
|- | |- | ||
|IS<i>6110</i>||Copy-paste circles||Rv2280 and PE-PGRS gene, Rv1468c||<i>Mycobacterium tuberculosis</i>||<ref><nowiki><pubmed>15130120</pubmed></nowiki></ref>||E | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS6110 IS<i>6110</i>]||Copy-paste circles||Rv2280 and PE-PGRS gene, Rv1468c||<i>Mycobacterium tuberculosis</i>||<ref><nowiki><pubmed>15130120</pubmed></nowiki></ref>||E | ||
|- | |- | ||
|IS<i>Kpn8</i>||Copy-paste circles||<i>blaKCP-2</i>||<i>Escherichia coli</i>, <i>Citrobacter freundii</i>, <i>Enterobacter cloacae</i>, <i>Enterobacter aerogenes</i>, and <i>Klebsiella oxytoca||<ref><nowiki><pubmed>24433026</pubmed></nowiki></ref>||C | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISKpn8 IS<i>Kpn8</i>]||Copy-paste circles||<i>blaKCP-2</i>||<i>Escherichia coli</i>, <i>Citrobacter freundii</i>, <i>Enterobacter cloacae</i>, <i>Enterobacter aerogenes</i>, and <i>Klebsiella oxytoca||<ref><nowiki><pubmed>24433026</pubmed></nowiki></ref>||C | ||
|- | |- | ||
| rowspan="9" |[[IS Families/IS4 and related families|IS<i>4</i>]]|| rowspan="3" |IS<i>10</i>||Cut-paste (hairpin)||<i>his</i>||<i>Salmonella typhimurium</i>||<ref><nowiki><pubmed>6289329</pubmed></nowiki></ref>||E | | rowspan="9" |[[IS Families/IS4 and related families|IS<i>4</i>]]|| rowspan="3" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10 IS<i>10</i>]||Cut-paste (hairpin)||<i>his</i>||<i>Salmonella typhimurium</i>||<ref><nowiki><pubmed>6289329</pubmed></nowiki></ref>||E | ||
|- | |- | ||
|Cut-paste||—||<i>Escherichia coli</i>||<ref><nowiki><pubmed>6311437</pubmed></nowiki></ref>||E | |Cut-paste||—||<i>Escherichia coli</i>||<ref><nowiki><pubmed>6311437</pubmed></nowiki></ref>||E | ||
| Line 55: | Line 55: | ||
|Cut-paste||<i>acrEF</i> pump||<i>Salmonella enterica</i>||<ref name=":6" />||E | |Cut-paste||<i>acrEF</i> pump||<i>Salmonella enterica</i>||<ref name=":6" />||E | ||
|- | |- | ||
|IS<i>50</i>||Cut-paste (hairpin)||<i>aph3’II</i>||<i>Escherichia coli</i>||<ref><nowiki><pubmed>6260374</pubmed></nowiki></ref>||E | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50 IS<i>50</i>]||Cut-paste (hairpin)||<i>aph3’II</i>||<i>Escherichia coli</i>||<ref><nowiki><pubmed>6260374</pubmed></nowiki></ref>||E | ||
|- | |- | ||
| rowspan="2" |IS<i>1999</i>|| rowspan="2" |—||<i>bla<sub>VEB-1</sub></i>||<i>Pseudomonas aeruginosa</i>||<ref name=":2" />||C | | rowspan="2" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1999 IS<i>1999</i>]|| rowspan="2" |—||<i>bla<sub>VEB-1</sub></i>||<i>Pseudomonas aeruginosa</i>||<ref name=":2" />||C | ||
|- | |- | ||
|<i>bla<sub>VEB-1</sub></i>/<i>bla<sub>OXA-48</sub></i>||<i>Escherichia coli</i>||<ref name=":1" />||C | |<i>bla<sub>VEB-1</sub></i>/<i>bla<sub>OXA-48</sub></i>||<i>Escherichia coli</i>||<ref name=":1" />||C | ||
|- | |- | ||
|IS<i>Pa12</i>||—||<i>blaPER-1</i>||<i>Salmonella enterica</i>, <i>Pseudomonas aeruginosa</i>, <i>Providencia stuartii</i>, <i>Acinetobacter baumannii</i>||<ref name=":9"><pubmed>12936998</pubmed></nowiki></ref>||C | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPa12 IS<i>Pa12</i>]||—||<i>blaPER-1</i>||<i>Salmonella enterica</i>, <i>Pseudomonas aeruginosa</i>, <i>Providencia stuartii</i>, <i>Acinetobacter baumannii</i>||<ref name=":9"><pubmed>12936998</pubmed></nowiki></ref>||C | ||
|- | |- | ||
| rowspan="2" |IS<i>Aba1</i>|| rowspan="2" |—||<i>bla<sub>ampC</sub></i>|| rowspan="2" |<i>Acinetobacter baumannii</i>||<ref name=":10"><pubmed>12951337</pubmed></nowiki></ref><ref><nowiki><pubmed>14742218</pubmed></nowiki></ref><ref><nowiki><pubmed>16441449</pubmed></nowiki></ref>||C | | rowspan="2" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAba1 IS<i>Aba1</i>]|| rowspan="2" |—||<i>bla<sub>ampC</sub></i>|| rowspan="2" |<i>Acinetobacter baumannii</i>||<ref name=":10"><pubmed>12951337</pubmed></nowiki></ref><ref><nowiki><pubmed>14742218</pubmed></nowiki></ref><ref><nowiki><pubmed>16441449</pubmed></nowiki></ref>||C | ||
|- | |- | ||
|<i>bla<sub>OXA-51</sub></i>/<i>bla<sub>OXA-23</sub></i>||<ref><nowiki><pubmed>16630258</pubmed></nowiki></ref>||C | |<i>bla<sub>OXA-51</sub></i>/<i>bla<sub>OXA-23</sub></i>||<ref><nowiki><pubmed>16630258</pubmed></nowiki></ref>||C | ||
|- | |- | ||
| rowspan="6" |[[IS Families/IS5 and related IS1182 families|IS<i>5</i>]] | | rowspan="6" |[[IS Families/IS5 and related IS1182 families|IS<i>5</i>]] | ||
|IS<i>5</i>|| rowspan="6" |—||<i>EmR</i>||<i>Escherichia coli</i>||<ref name=":8" />||C? | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS<i>5</i>]|| rowspan="6" |—||<i>EmR</i>||<i>Escherichia coli</i>||<ref name=":8" />||C? | ||
|- | |- | ||
|ISFtu2||general||<i>Francisella tularensis</i>||<ref name=":11"><pubmed>19749055</pubmed></nowiki></ref>||Natural isolate | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISFtu2 IS''Ftu2'']||general||<i>Francisella tularensis</i>||<ref name=":11"><pubmed>19749055</pubmed></nowiki></ref>||Natural isolate | ||
|- | |- | ||
|ISVa1||(iron uptake)||<i>Vibrio anguilarum</i>||<ref><nowiki><pubmed>7568465</pubmed></nowiki></ref>||Natural isolate | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISVa1 IS''Va1'']||(iron uptake)||<i>Vibrio anguilarum</i>||<ref><nowiki><pubmed>7568465</pubmed></nowiki></ref>||Natural isolate | ||
|- | |- | ||
|IS<i>1168</i> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1168 IS<i>1168</i>] ||<i>nimA</i>, <i>nimB</i>||<i>Bacteroides</i> sp.||<ref><nowiki><pubmed>8067736</pubmed></nowiki></ref>||C | ||
|- | |- | ||
|IS<i>1186</i>||<i>cfiA</i>||<i>Bacteroides fragilis</i>||<ref name=":12"><pubmed>8057831</pubmed></nowiki></ref><ref><nowiki><pubmed>7545155</pubmed></nowiki></ref>||C | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1186 IS<i>1186</i>]||<i>cfiA</i>||<i>Bacteroides fragilis</i>||<ref name=":12"><pubmed>8057831</pubmed></nowiki></ref><ref><nowiki><pubmed>7545155</pubmed></nowiki></ref>||C | ||
|- | |- | ||
|IS<i>402</i>||<i>bla</i>||<i>Pseudomonas cepacia</i>||<ref name=":13"><pubmed>3025189</pubmed></nowiki></ref>||E | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS402 IS<i>402</i>]||<i>bla</i>||<i>Pseudomonas cepacia</i>||<ref name=":13"><pubmed>3025189</pubmed></nowiki></ref>||E | ||
|- | |- | ||
| rowspan="6" |[[IS Families/IS6 family|IS<i>6</i>]] | | rowspan="6" |[[IS Families/IS6 family|IS<i>6</i>]] | ||
|IS<i>257</i>||Cointegrate||<i>dfrA</i>||<i>Staphylococcus aureus</i>||<ref><nowiki><pubmed>7840551</pubmed></nowiki></ref>|| rowspan="6" |C | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS257 IS<i>257</i>]||Cointegrate||<i>dfrA</i>||<i>Staphylococcus aureus</i>||<ref><nowiki><pubmed>7840551</pubmed></nowiki></ref>|| rowspan="6" |C | ||
|- | |- | ||
|IS<i>257</i>||Cointegrate||<i>tet</i>||<i>Staphylococcus aureus</i>||<ref><nowiki><pubmed>10852863</pubmed></nowiki></ref> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS257 IS<i>257</i>]||Cointegrate||<i>tet</i>||<i>Staphylococcus aureus</i>||<ref><nowiki><pubmed>10852863</pubmed></nowiki></ref> | ||
|- | |- | ||
|IS<i>1008</i>||—||<i>bla<sub>OXA-58</sub></i>||<i>Acinetobacter baumannii</i>||<ref><nowiki><pubmed>18443121</pubmed></nowiki></ref> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1008 IS<i>1008</i>]||—||<i>bla<sub>OXA-58</sub></i>||<i>Acinetobacter baumannii</i>||<ref><nowiki><pubmed>18443121</pubmed></nowiki></ref> | ||
|- | |- | ||
| rowspan="2" |IS<i>26</i>|| rowspan="2" |—||<i>aphA7</i>, <i>bla<sub>S2A</sub></i>||<i>Klebsiella pneumoniae</i>||<ref><nowiki><pubmed>2160941</pubmed></nowiki></ref> | | rowspan="2" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS26 IS<i>26</i>]|| rowspan="2" |—||<i>aphA7</i>, <i>bla<sub>S2A</sub></i>||<i>Klebsiella pneumoniae</i>||<ref><nowiki><pubmed>2160941</pubmed></nowiki></ref> | ||
|- | |- | ||
|<i>bla<sub>SHV-2a</sub></i>||<i>Pseudomonas aeruginosa</i>||<ref><nowiki><pubmed>10223953</pubmed></nowiki></ref> | |<i>bla<sub>SHV-2a</sub></i>||<i>Pseudomonas aeruginosa</i>||<ref><nowiki><pubmed>10223953</pubmed></nowiki></ref> | ||
|- | |- | ||
|IS<i>140</i>||—||<i>aac(3)</i>-III and -IV||—||<ref><nowiki><pubmed>6318050</pubmed></nowiki></ref> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS140 IS<i>140</i>]||—||<i>aac(3)</i>-III and -IV||—||<ref><nowiki><pubmed>6318050</pubmed></nowiki></ref> | ||
|- | |- | ||
| rowspan="2" |[[IS Families/IS21 family|IS<i>21</i>]] | | rowspan="2" |[[IS Families/IS21 family|IS<i>21</i>]] | ||
|IS<i>Bf1</i>|| rowspan="2" |Copy-paste circle||<i>cepA</i>||<i>Bacteroides fragilis</i>||<ref><nowiki><pubmed>7517394</pubmed></nowiki></ref>|| rowspan="2" |C | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISBf1 IS<i>Bf1</i>]|| rowspan="2" |Copy-paste circle||<i>cepA</i>||<i>Bacteroides fragilis</i>||<ref><nowiki><pubmed>7517394</pubmed></nowiki></ref>|| rowspan="2" |C | ||
|- | |- | ||
|IS<i>Kpn7</i>||<i>blaKPC</i>||<i>Klebsiellea pneumonia</i>||<ref><nowiki><pubmed>18227185</pubmed></nowiki></ref> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISKpn7 IS<i>Kpn7</i>]||<i>blaKPC</i>||<i>Klebsiellea pneumonia</i>||<ref><nowiki><pubmed>18227185</pubmed></nowiki></ref> | ||
|- | |- | ||
| rowspan="6" |[[IS Families/IS30 family|IS<i>30</i>]] | | rowspan="6" |[[IS Families/IS30 family|IS<i>30</i>]] | ||
|IS<i>30</i>|| rowspan="6" |Copy-paste circle||<i>galK</i>||<i>Escherichia coli</i>||<ref><nowiki><pubmed>3039299</pubmed></nowiki></ref>||E | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS30 IS<i>30</i>]|| rowspan="6" |Copy-paste circle||<i>galK</i>||<i>Escherichia coli</i>||<ref><nowiki><pubmed>3039299</pubmed></nowiki></ref>||E | ||
|- | |- | ||
| rowspan="2" |IS<i>18</i>||<i>aac(6’)-Ij</i>|| rowspan="2" |<i>Acinetobacter</i> sp.||<ref><nowiki><pubmed>9756793</pubmed></nowiki></ref>|| rowspan="2" |C | | rowspan="2" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS18 IS<i>18</i>]||<i>aac(6’)-Ij</i>|| rowspan="2" |<i>Acinetobacter</i> sp.||<ref><nowiki><pubmed>9756793</pubmed></nowiki></ref>|| rowspan="2" |C | ||
|- | |- | ||
|<i>blaOXA257</i>||<ref><nowiki><pubmed>23014718</pubmed></nowiki></ref> | |<i>blaOXA257</i>||<ref><nowiki><pubmed>23014718</pubmed></nowiki></ref> | ||
|- | |- | ||
| rowspan="2" |IS<i>4351</i>||<i>ermF</i>|| rowspan="2" |<i>Bacteroides fragilis</i>||<ref><nowiki><pubmed>3038844</pubmed></nowiki></ref>|| rowspan="2" |C | | rowspan="2" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS4351 IS<i>4351</i>]||<i>ermF</i>|| rowspan="2" |<i>Bacteroides fragilis</i>||<ref><nowiki><pubmed>3038844</pubmed></nowiki></ref>|| rowspan="2" |C | ||
|- | |- | ||
|<i>cfiA</i>||<ref name=":3" /> | |<i>cfiA</i>||<ref name=":3" /> | ||
|- | |- | ||
|IS<i>1086</i>||<i>cnrCBAT</i> (Zn<sup>R</sup>)||<i>Cupriavidus metallidurans</i>|| ||E | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1086 IS<i>1086</i>]||<i>cnrCBAT</i> (Zn<sup>R</sup>)||<i>Cupriavidus metallidurans</i>|| ||E | ||
|- | |- | ||
| rowspan="4" |[[IS Families/IS256 family|IS<i>256</i>]] | | rowspan="4" |[[IS Families/IS256 family|IS<i>256</i>]] | ||
| rowspan="2" |IS<i>256</i>|| rowspan="4" |Copy-paste circles||<i>mecA</i>||<i>Staphylococcus sciuri</i>|| rowspan="2" |<ref><nowiki><pubmed>9371438</pubmed></nowiki></ref><ref><nowiki><pubmed>12511511</pubmed></nowiki></ref>|| rowspan="2" |— | | rowspan="2" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS256 IS<i>256</i>]|| rowspan="4" |Copy-paste circles||<i>mecA</i>||<i>Staphylococcus sciuri</i>|| rowspan="2" |<ref><nowiki><pubmed>9371438</pubmed></nowiki></ref><ref><nowiki><pubmed>12511511</pubmed></nowiki></ref>|| rowspan="2" |— | ||
|- | |- | ||
|<i>llm</i>||<i>Staphylococcus aureus</i> | |<i>llm</i>||<i>Staphylococcus aureus</i> | ||
|- | |- | ||
|IS<i>1490</i>||—||<i>Burkholderia cepacia</i>||<ref><nowiki><pubmed>9098071</pubmed></nowiki></ref>||— | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1490 IS<i>1490</i>]||—||<i>Burkholderia cepacia</i>||<ref><nowiki><pubmed>9098071</pubmed></nowiki></ref>||— | ||
|- | |- | ||
|IS<i>406</i>||—||<i>Burkholderia cepacia</i>||<ref name=":13" />||E | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS406 IS<i>406</i>]||—||<i>Burkholderia cepacia</i>||<ref name=":13" />||E | ||
|- | |- | ||
| rowspan="2" |[[IS Families/IS481 family|IS<i>481</i>]]||IS<i>481</i>|| rowspan="2" |Copy-paste circles||<i>katA</i>||<i>Bordetella pertussis</i>||<ref><nowiki><pubmed>7830550</pubmed></nowiki></ref>||— | | rowspan="2" |[[IS Families/IS481 family|IS<i>481</i>]]||[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS481 IS<i>481</i>]|| rowspan="2" |Copy-paste circles||<i>katA</i>||<i>Bordetella pertussis</i>||<ref><nowiki><pubmed>7830550</pubmed></nowiki></ref>||— | ||
|- | |- | ||
|IS<i>Rme5</i>||<i>cnrCBAT</i> (Zn R)||<i>Cupriavidus metallidurans</i>||<ref><nowiki><pubmed>27047473</pubmed></nowiki></ref>||E | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISRme5 IS<i>Rme5</i>]||<i>cnrCBAT</i> (Zn R)||<i>Cupriavidus metallidurans</i>||<ref><nowiki><pubmed>27047473</pubmed></nowiki></ref>||E | ||
|- | |- | ||
|[[IS Families/IS630 family|IS<i>630</i>]]||IS<i>Ftu1</i>||Tc-like||general||<i>Francisella tularensis</i>||<ref name=":11" />||Natural isolate | |[[IS Families/IS630 family|IS<i>630</i>]]||[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISFtu1 IS<i>Ftu1</i>]||Tc-like||general||<i>Francisella tularensis</i>||<ref name=":11" />||Natural isolate | ||
|- | |- | ||
| rowspan="3" |[[IS Families/IS982 family|IS<i>982</i>]] | | rowspan="3" |[[IS Families/IS982 family|IS<i>982</i>]] | ||
| rowspan="2" |IS<i>1187</i>|| rowspan="3" |—|| rowspan="2" |<i>cfiA</i>|| rowspan="2" |<i>Bacteroides fragilis</i>||<ref name=":12" />||E+C | | rowspan="2" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1187 IS<i>1187</i>]|| rowspan="3" |—|| rowspan="2" |<i>cfiA</i>|| rowspan="2" |<i>Bacteroides fragilis</i>||<ref name=":12" />||E+C | ||
|- | |- | ||
|<ref name=":3" /><ref name=":14"><pubmed>11344163</pubmed></nowiki></ref><ref name=":15"><pubmed>12604530</pubmed></nowiki></ref>||C | |<ref name=":3" /><ref name=":14"><pubmed>11344163</pubmed></nowiki></ref><ref name=":15"><pubmed>12604530</pubmed></nowiki></ref>||C | ||
|- | |- | ||
|IS<i>982</i>||<i>citQRP</i>||<i>Lactococcus lactis</i>||<ref><nowiki><pubmed>8602160</pubmed></nowiki></ref>||— | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS982 IS<i>982</i>]||<i>citQRP</i>||<i>Lactococcus lactis</i>||<ref><nowiki><pubmed>8602160</pubmed></nowiki></ref>||— | ||
|- | |- | ||
| rowspan="12" |[[IS Families/IS1380 family|IS<i>1380</i>]] | | rowspan="12" |[[IS Families/IS1380 family|IS<i>1380</i>]] | ||
|IS<i>Bf12</i>|| rowspan="12" |—|| rowspan="6" |<i>cfiA</i>|| rowspan="6" |<i>Bacteroides fragilis</i>||<ref name=":3" />|| rowspan="12" |C | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISBf12 IS<i>Bf12</i>]|| rowspan="12" |—|| rowspan="6" |<i>cfiA</i>|| rowspan="6" |<i>Bacteroides fragilis</i>||<ref name=":3" />|| rowspan="12" |C | ||
|- | |- | ||
|IS<i>612</i>||<ref name=":15" /> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS612 IS<i>612</i>]||<ref name=":15" /> | ||
|- | |- | ||
|IS<i>613</i>||<ref name=":15" /> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS613 IS<i>613</i>]||<ref name=":15" /> | ||
|- | |- | ||
|IS<i>614</i>||<ref name=":3" /><ref name=":15" /><ref><nowiki><pubmed>19744834</pubmed></nowiki></ref> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS614 IS<i>614</i>]||<ref name=":3" /><ref name=":15" /><ref><nowiki><pubmed>19744834</pubmed></nowiki></ref> | ||
|- | |- | ||
|IS<i>1188</i>||<ref name=":14" /> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1188 IS<i>1188</i>]||<ref name=":14" /> | ||
|- | |- | ||
|IS<i>942</i>||<ref name=":14" /> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS942 IS<i>942</i>]||<ref name=":14" /> | ||
|- | |- | ||
| rowspan="5" |IS<i>Ecp1</i>||<i>bla<sub>CTX-M-15</sub></i>||Enterobacteriaceae||<ref><nowiki><pubmed>11470367</pubmed></nowiki></ref> | | rowspan="5" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS<i>Ecp1</i>]||<i>bla<sub>CTX-M-15</sub></i>||Enterobacteriaceae||<ref><nowiki><pubmed>11470367</pubmed></nowiki></ref> | ||
|- | |- | ||
|<i>bla<sub>CTX-M-17</sub></i>||<i>Klebsiella pneumoniae</i>||<ref><nowiki><pubmed>12435670</pubmed></nowiki></ref> | |<i>bla<sub>CTX-M-17</sub></i>||<i>Klebsiella pneumoniae</i>||<ref><nowiki><pubmed>12435670</pubmed></nowiki></ref> | ||
| Line 156: | Line 156: | ||
|<i>rmtC</i>||<i>Escherichia coli</i>||<ref><nowiki><pubmed>16940134</pubmed></nowiki></ref> | |<i>rmtC</i>||<i>Escherichia coli</i>||<ref><nowiki><pubmed>16940134</pubmed></nowiki></ref> | ||
|- | |- | ||
|IS<i>1187</i>||<i>cfiA</i>||<i>Bacteroides fragilis</i>||<ref name=":14" /> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1187 IS<i>1187</i>]||<i>cfiA</i>||<i>Bacteroides fragilis</i>||<ref name=":14" /> | ||
|- | |- | ||
| rowspan="2" |[[IS Families/ISL3 family|IS<i>L3</i>]] | | rowspan="2" |[[IS Families/ISL3 family|IS<i>L3</i>]] | ||
|IS<i>Sg1</i>|| rowspan="2" |Copy-paste circles||<i>sspB</i> (surface antigen)||<i>Streptococcus gordonii</i>||<ref><nowiki><pubmed>9202480</pubmed></nowiki></ref>|| rowspan="2" |— | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISSg1 IS<i>Sg1</i>]|| rowspan="2" |Copy-paste circles||<i>sspB</i> (surface antigen)||<i>Streptococcus gordonii</i>||<ref><nowiki><pubmed>9202480</pubmed></nowiki></ref>|| rowspan="2" |— | ||
|- | |- | ||
|IS<i>1411</i>||<i>pheBA</i>||<i>Pseudomonas putida</i>||<ref><nowiki><pubmed>9765560</pubmed></nowiki></ref> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1411 IS<i>1411</i>]||<i>pheBA</i>||<i>Pseudomonas putida</i>||<ref><nowiki><pubmed>9765560</pubmed></nowiki></ref> | ||
|- | |- | ||
| rowspan="2" |[[IS Families/ISAs1 family|IS<i>As1</i>]] | | rowspan="2" |[[IS Families/ISAs1 family|IS<i>As1</i>]] | ||
|IS<i>1548</i>|| rowspan="2" |—||<i>lmb</i> (lamelin binding)||<i>Streptococcus agalactiae</i>||<ref><nowiki><pubmed>20520730</pubmed></nowiki></ref>||— | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1548 IS<i>1548</i>]|| rowspan="2" |—||<i>lmb</i> (lamelin binding)||<i>Streptococcus agalactiae</i>||<ref><nowiki><pubmed>20520730</pubmed></nowiki></ref>||— | ||
|- | |- | ||
|nd||—||<i>Acinetobacter baumannii</i>||<ref name=":10" />||C | |nd||—||<i>Acinetobacter baumannii</i>||<ref name=":10" />||C | ||
|- | |- | ||
| rowspan="3" |IS<i>NYC</i> | | rowspan="3" |IS<i>NYC</i> | ||
|IS<i>403</i>|| rowspan="3" |—|| rowspan="3" |bla|| rowspan="3" |<i>Burkholderia cepacia</i>|| rowspan="3" |<ref name=":13" />|| rowspan="3" |E | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS403 IS<i>403</i>]|| rowspan="3" |—|| rowspan="3" |''bla''|| rowspan="3" |<i>Burkholderia cepacia</i>|| rowspan="3" |<ref name=":13" />|| rowspan="3" |E | ||
|- | |- | ||
|IS<i>404</i> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS404 IS<i>404</i>] | ||
|- | |- | ||
|IS<i>405</i> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS405 IS<i>405</i>] | ||
|} | |} | ||
Revision as of 09:10, 9 August 2021
Another important aspect of IS impact on their bacterial hosts is their ability to modulate gene expression. In addition to acting as vectors for gene transmission from one replicon to another in the form composite transposons (two IS flanking any gene; Fig.2.3) and tIS (Fig.8.1) and their ability to interrupt genes, it has been known for some time[1][2] that IS can also activate gene expression. This capacity has recently received much attention due to the increase in resistance to various antibacterials[3][4][5], a worrying public health threat[6][7].
They can accomplish this in two ways: either by providing internal promoters whose transcripts escape into neighboring DNA[2][8][9][10] or by hybrid promoter formation. Many IS carry -35 promoter components oriented towards the flanking DNA (Fig.18.1). In a number of cases this plays an important part in their transposition since a significant number of IS transposes using an excised transposon circle (Fig.18.1) with abutted left and right ends. For these IS, the other end carries a -10 element oriented inwards towards the Tpase gene. Together with the -35, this generates a strong promoter on formation of the circle junction to drive Tpase expression required for catalysis of integration (Fig.18.2) [11][12][13][14]. Thus if integration occurs next to a resident -10 sequence, the IS -35 sequence can contribute to a hybrid promoter to drive expression of neighboring genes [see [15]]. At present this phenomenon had been reported to occur with over 30 different IS in more than 17 bacterial species[16][17] (Table IS and Gene Expression below). Indeed, specific vector plasmids have been designed to identify activating insertions (e.g. [18]).
IS activity can affect efflux mechanisms resulting in increased resistance: IS1 or IS10 insertion can up-regulate the AcrAB-TolC pump in Salmonella enterica[19]; IS1 or IS2 insertion upstream of AcrEF[20][21] and IS186 insertional inactivation of the AcrAB repressor, AcrR, in Escherichia coli [20], all lead to increased resistance to fluoroquinolones. Insertional inactivation of specific porins can also play a significant role[22].
IS and Gene Expression
| Table. IS and gene expression. | ||||||||
| IS family | IS name | Mechanism | Gene(s) affected | Organism | Reference | Clinical/Experimental | ||
|---|---|---|---|---|---|---|---|---|
| IS1 | IS1 | Cointegrate | EmR | Escherichia coli | [23] | C? | ||
| Copy-paste circles | blaTEM-1 | [15] | E | |||||
| — | acrEF pump | Salmonella enterica | [19] | E | ||||
| IS3 | IS2 | Copy-paste circles | gal | Escherichia coli | [24][25] | E | ||
| gal | [23][2] | E | ||||||
| argE | [26][20] | E | ||||||
| EmR | C? | |||||||
| blaampC | E | |||||||
| acrEF pump | E | |||||||
| IS3 | Copy-paste circles | argE | Escherichia coli | [2] | E | |||
| argE | [8] | E | ||||||
| citT | [27] | E | ||||||
| IS981 | Copy-paste circles | ldhB | Lactococcus lactis | [28] | E | |||
| IS6110 | Copy-paste circles | Rv2280 and PE-PGRS gene, Rv1468c | Mycobacterium tuberculosis | [29] | E | |||
| ISKpn8 | Copy-paste circles | blaKCP-2 | Escherichia coli, Citrobacter freundii, Enterobacter cloacae, Enterobacter aerogenes, and Klebsiella oxytoca | [30] | C | |||
| IS4 | IS10 | Cut-paste (hairpin) | his | Salmonella typhimurium | [31] | E | ||
| Cut-paste | — | Escherichia coli | [32] | E | ||||
| Cut-paste | acrEF pump | Salmonella enterica | [19] | E | ||||
| IS50 | Cut-paste (hairpin) | aph3’II | Escherichia coli | [33] | E | |||
| IS1999 | — | blaVEB-1 | Pseudomonas aeruginosa | [4] | C | |||
| blaVEB-1/blaOXA-48 | Escherichia coli | [3] | C | |||||
| ISPa12 | — | blaPER-1 | Salmonella enterica, Pseudomonas aeruginosa, Providencia stuartii, Acinetobacter baumannii | [34] | C | |||
| ISAba1 | — | blaampC | Acinetobacter baumannii | [35][36][37] | C | |||
| blaOXA-51/blaOXA-23 | [38] | C | ||||||
| IS5 | IS5 | — | EmR | Escherichia coli | [23] | C? | ||
| ISFtu2 | general | Francisella tularensis | [39] | Natural isolate | ||||
| ISVa1 | (iron uptake) | Vibrio anguilarum | [40] | Natural isolate | ||||
| IS1168 | nimA, nimB | Bacteroides sp. | [41] | C | ||||
| IS1186 | cfiA | Bacteroides fragilis | [42][43] | C | ||||
| IS402 | bla | Pseudomonas cepacia | [44] | E | ||||
| IS6 | IS257 | Cointegrate | dfrA | Staphylococcus aureus | [45] | C | ||
| IS257 | Cointegrate | tet | Staphylococcus aureus | [46] | ||||
| IS1008 | — | blaOXA-58 | Acinetobacter baumannii | [47] | ||||
| IS26 | — | aphA7, blaS2A | Klebsiella pneumoniae | [48] | ||||
| blaSHV-2a | Pseudomonas aeruginosa | [49] | ||||||
| IS140 | — | aac(3)-III and -IV | — | [50] | ||||
| IS21 | ISBf1 | Copy-paste circle | cepA | Bacteroides fragilis | [51] | C | ||
| ISKpn7 | blaKPC | Klebsiellea pneumonia | [52] | |||||
| IS30 | IS30 | Copy-paste circle | galK | Escherichia coli | [53] | E | ||
| IS18 | aac(6’)-Ij | Acinetobacter sp. | [54] | C | ||||
| blaOXA257 | [55] | |||||||
| IS4351 | ermF | Bacteroides fragilis | [56] | C | ||||
| cfiA | [5] | |||||||
| IS1086 | cnrCBAT (ZnR) | Cupriavidus metallidurans | E | |||||
| IS256 | IS256 | Copy-paste circles | mecA | Staphylococcus sciuri | [57][58] | — | ||
| llm | Staphylococcus aureus | |||||||
| IS1490 | — | Burkholderia cepacia | [59] | — | ||||
| IS406 | — | Burkholderia cepacia | [44] | E | ||||
| IS481 | IS481 | Copy-paste circles | katA | Bordetella pertussis | [60] | — | ||
| ISRme5 | cnrCBAT (Zn R) | Cupriavidus metallidurans | [61] | E | ||||
| IS630 | ISFtu1 | Tc-like | general | Francisella tularensis | [39] | Natural isolate | ||
| IS982 | IS1187 | — | cfiA | Bacteroides fragilis | [42] | E+C | ||
| [5][62][63] | C | |||||||
| IS982 | citQRP | Lactococcus lactis | [64] | — | ||||
| IS1380 | ISBf12 | — | cfiA | Bacteroides fragilis | [5] | C | ||
| IS612 | [63] | |||||||
| IS613 | [63] | |||||||
| IS614 | [5][63][65] | |||||||
| IS1188 | [62] | |||||||
| IS942 | [62] | |||||||
| ISEcp1 | blaCTX-M-15 | Enterobacteriaceae | [66] | |||||
| blaCTX-M-17 | Klebsiella pneumoniae | [67] | ||||||
| blaCTX-M-19 | Klebsiella pneumoniae | [34] | ||||||
| blaCTX-M | Kluyvera ascorbata | [68] | ||||||
| rmtC | Escherichia coli | [69] | ||||||
| IS1187 | cfiA | Bacteroides fragilis | [62] | |||||
| ISL3 | ISSg1 | Copy-paste circles | sspB (surface antigen) | Streptococcus gordonii | [70] | — | ||
| IS1411 | pheBA | Pseudomonas putida | [71] | |||||
| ISAs1 | IS1548 | — | lmb (lamelin binding) | Streptococcus agalactiae | [72] | — | ||
| nd | — | Acinetobacter baumannii | [35] | C | ||||
| ISNYC | IS403 | — | bla | Burkholderia cepacia | [44] | E | ||
| IS404 | ||||||||
| IS405 | ||||||||
Bibliography
- ↑ <pubmed>1101028</pubmed>
- ↑ 2.0 2.1 2.2 2.3 <pubmed>6271458</pubmed></nowiki>
- ↑ 3.0 3.1 <pubmed>16952941</pubmed></nowiki>
- ↑ 4.0 4.1 <pubmed>12923109</pubmed></nowiki>
- ↑ 5.0 5.1 5.2 5.3 5.4 <pubmed>23158541</pubmed></nowiki>
- ↑ <pubmed>23887414</pubmed>
- ↑ <pubmed>23887415</pubmed>
- ↑ 8.0 8.1 <pubmed>6292860</pubmed></nowiki>
- ↑ <pubmed>6260746</pubmed>
- ↑ <pubmed>6311437</pubmed>
- ↑ <pubmed>26350305</pubmed>
- ↑ <pubmed>9214651</pubmed>
- ↑ <pubmed>10438765</pubmed>
- ↑ <pubmed>11598022</pubmed>
- ↑ 15.0 15.1 <pubmed>3029382</pubmed></nowiki>
- ↑ <pubmed>17223624</pubmed>
- ↑ <pubmed>24499397</pubmed>
- ↑ <pubmed>8863735</pubmed>
- ↑ 19.0 19.1 19.2 <pubmed>15616308</pubmed></nowiki>
- ↑ 20.0 20.1 20.2 <pubmed>11302812</pubmed></nowiki>
- ↑ <pubmed>11274125</pubmed>
- ↑ <pubmed>15212803</pubmed>
- ↑ 23.0 23.1 23.2 <pubmed>6283551</pubmed></nowiki>
- ↑ <pubmed>4610339</pubmed>
- ↑ <pubmed>339095</pubmed>
- ↑ <pubmed>6187472</pubmed>
- ↑ <pubmed>22992527</pubmed>
- ↑ <pubmed>12867459</pubmed>
- ↑ <pubmed>15130120</pubmed>
- ↑ <pubmed>24433026</pubmed>
- ↑ <pubmed>6289329</pubmed>
- ↑ <pubmed>6311437</pubmed>
- ↑ <pubmed>6260374</pubmed>
- ↑ 34.0 34.1 <pubmed>12936998</pubmed></nowiki>
- ↑ 35.0 35.1 <pubmed>12951337</pubmed></nowiki>
- ↑ <pubmed>14742218</pubmed>
- ↑ <pubmed>16441449</pubmed>
- ↑ <pubmed>16630258</pubmed>
- ↑ 39.0 39.1 <pubmed>19749055</pubmed></nowiki>
- ↑ <pubmed>7568465</pubmed>
- ↑ <pubmed>8067736</pubmed>
- ↑ 42.0 42.1 <pubmed>8057831</pubmed></nowiki>
- ↑ <pubmed>7545155</pubmed>
- ↑ 44.0 44.1 44.2 <pubmed>3025189</pubmed></nowiki>
- ↑ <pubmed>7840551</pubmed>
- ↑ <pubmed>10852863</pubmed>
- ↑ <pubmed>18443121</pubmed>
- ↑ <pubmed>2160941</pubmed>
- ↑ <pubmed>10223953</pubmed>
- ↑ <pubmed>6318050</pubmed>
- ↑ <pubmed>7517394</pubmed>
- ↑ <pubmed>18227185</pubmed>
- ↑ <pubmed>3039299</pubmed>
- ↑ <pubmed>9756793</pubmed>
- ↑ <pubmed>23014718</pubmed>
- ↑ <pubmed>3038844</pubmed>
- ↑ <pubmed>9371438</pubmed>
- ↑ <pubmed>12511511</pubmed>
- ↑ <pubmed>9098071</pubmed>
- ↑ <pubmed>7830550</pubmed>
- ↑ <pubmed>27047473</pubmed>
- ↑ 62.0 62.1 62.2 62.3 <pubmed>11344163</pubmed></nowiki>
- ↑ 63.0 63.1 63.2 63.3 <pubmed>12604530</pubmed></nowiki>
- ↑ <pubmed>8602160</pubmed>
- ↑ <pubmed>19744834</pubmed>
- ↑ <pubmed>11470367</pubmed>
- ↑ <pubmed>12435670</pubmed>
- ↑ <pubmed>16569841</pubmed>
- ↑ <pubmed>16940134</pubmed>
- ↑ <pubmed>9202480</pubmed>
- ↑ <pubmed>9765560</pubmed>
- ↑ <pubmed>20520730</pubmed>