IS Families/IS200 IS605 family: Difference between revisions
No edit summary |
No edit summary |
||
| Line 399: | Line 399: | ||
This is similar to results obtained with Cas9 and Cas12 systems themselves <ref><nowiki><pubmed>23287718</pubmed></nowiki></ref><ref><nowiki><pubmed>26422227</pubmed></nowiki></ref>. Finally, variation in 5’ length showed that shortest active scaffolds were 120–140 nt long and lengths of 300 nts were active. | This is similar to results obtained with Cas9 and Cas12 systems themselves <ref><nowiki><pubmed>23287718</pubmed></nowiki></ref><ref><nowiki><pubmed>26422227</pubmed></nowiki></ref>. Finally, variation in 5’ length showed that shortest active scaffolds were 120–140 nt long and lengths of 300 nts were active. | ||
[[File:FigIS200 605 42.png|center|thumb|720x720px|'''Fig. IS200.42. Overall interactions between TnpB, reRNA and target DNA. i) Structure of the right end of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISDra2 IS''Dra2'']''' <ref | [[File:FigIS200 605 42.png|center|thumb|720x720px|'''Fig. IS200.42. Overall interactions between TnpB, reRNA and target DNA. i) Structure of the right end of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISDra2 IS''Dra2'']''' <ref name=":9" /> showing a cartoon of the secondary stricture, the DNA sequence from -30 to -1 and the base pairing observed between G<sub>R</sub> and C<sub>R</sub>. '''ii)''' '''reRNA from -119 to +16''' showing detailed secondary structures. Note that the colors are those shown in ('''iii'''). The guide sequence is shown in red. The G<sub>R</sub> and C<sub>R</sub> sequence equivalents in reRNA are boxed. '''iii)''' '''two dimentional representation of reRNA structures''' in the TnpB-RNP complex (left) and in the Ternary complex with target DNA (right). The dark green, yellow and grey circles surrounding each nucleotide indicate the interacting segments of TnpB (insert below). Note that in the target sequence, the 5 nucleotide sequence 3’ to TAM is shown as complementary, however, for technical reasons (to facilitate unpairing ready for interaction with the reRNA quide sequence), the sequence CTCAG was used <ref name=":33" />.]] | ||
For Dra2TnpB, the C-terminal domain (residues 376 to 408; [[:File:FigIS200 605 42.png|Fig. IS200.42]] '''bottom insert''') has relatively low sequence similarity among TnpB proteins and is disordered in the structures. The C-terminal truncation mutant (Δ376 to 408; ΔCTD) is efficient in target DNA cleavage but exhibits somewhat reduced protein stability. Thus the CTD is not required for RNA-guided target DNA cleavage. | For Dra2TnpB, the C-terminal domain (residues 376 to 408; [[:File:FigIS200 605 42.png|Fig. IS200.42]] '''bottom insert''') has relatively low sequence similarity among TnpB proteins and is disordered in the structures. The C-terminal truncation mutant (Δ376 to 408; ΔCTD) is efficient in target DNA cleavage but exhibits somewhat reduced protein stability. Thus the CTD is not required for RNA-guided target DNA cleavage. | ||
| Line 494: | Line 494: | ||
<br /> | <br /> | ||
==== '''The Structure of IsrB–ωRNA ribonucleoprotein complex and the ternary complex containing target DNA''' ==== | ===='''The Structure of IsrB–ωRNA ribonucleoprotein complex and the ternary complex containing target DNA'''==== | ||
IsrB is short, about 350 amino acids and lacking an [https://www.ebi.ac.uk/interpro/entry/InterPro/IPR003615/ HNH domain] ([[:File:FigIS200 605 51.png|Fig. IS200.51]]) (therefore equivalent to the ??HNH IscB derivative). It is associated with a long RNA guide of ~300-nt which guides IsrB to nick the non-target strand (NTS) of double-stranded (ds) DNA (see [[:File:FigIS200 605 51.png|Fig. IS200.51]] '''top''') containing a 5′-NTGA-3′ '''TAM <ref name=":30" />'''. | IsrB is short, about 350 amino acids and lacking an [https://www.ebi.ac.uk/interpro/entry/InterPro/IPR003615/ HNH domain] ([[:File:FigIS200 605 51.png|Fig. IS200.51]]) (therefore equivalent to the ??HNH IscB derivative). It is associated with a long RNA guide of ~300-nt which guides IsrB to nick the non-target strand (NTS) of double-stranded (ds) DNA (see [[:File:FigIS200 605 51.png|Fig. IS200.51]] '''top''') containing a 5′-NTGA-3′ '''TAM <ref name=":30" />'''. | ||
Revision as of 19:16, 29 June 2024
Historical
One of the founding members of this group, IS200, was identified in Salmonella typhimurium [1] as a mutation in hisD (hisD984) which mapped as a point mutation but which did not revert and was polar on the downstream hisC gene (see [2]). S. typhimurium LT2 was found to contain six IS200 copies and the IS was unique to Salmonella [3]. Further studies [4] showed that the IS did not carry repeated sequences, either direct or inverted, at its ends, and that removal of 50 bp at the transposase proximal end (which includes a structure resembling a transcription terminator) removed the strong transcriptional block. IS200 elements from S. typhimurium and S. abortusovis revealed a highly conserved structure of 707–708 bp with a single open-reading-frame potentially encoding a 151 aa peptide and a putative upstream ribosome-binding-site [5].
It has been suggested that a combination of inefficient transcription, protection from impinging transcription by a transcriptional terminator, and repression of translation by a stem-loop mRNA structure. All contribute to tight repression of transposase synthesis [2]. However, although IS200 seems to be relatively inactive in transposition [6], it is involved in chromosome arrangements in S. typhimurium by recombination between copies [7].
A second group of “founding” members of this family was, arguably, IS1341 from the thermophilic bacterium PS3 [8], IS891 from Anabaena sp. M-131 [9] and IS1136 from Saccharopolyspora erythraea [10]. The “transposases” of both elements were observed to be associated in a single IS, IS605, from the gastric pathogen Helicobacter pylori [11]. It was identified in many independent isolates of H. pylori and is now considered to be a central member which defines this large family. IS605 was shown to possess unique, not inverted repeat, ends; did not duplicate target sequences during transposition; and inserted with its left (IS200-homolog) end abutting 5'-TTTAA or 5'-TTTAAC target sequences [11]. Additionally, a second derivative, IS606, with only 25% amino acid identity in the two proteins (orfA and orfB) was also identified in many of the H. pylori isolates including some which were devoid of IS605. The Berg lab also identified another H. pylori IS, IS607 [12] which carried a similar IS1341-like orf (orfB) but with another upstream orf with similarities to that of the mycobacterial IS1535 [13] annotated as a resolvase due the presence of a site-specific serine recombinase motif. Another IS605 derivative, ISHp608, which appeared widely distributed in H. pylori was shown to transpose in E. coli, required only orfA to transpose and inserted downstream from a 5’-TTAC target sequence [14].
General
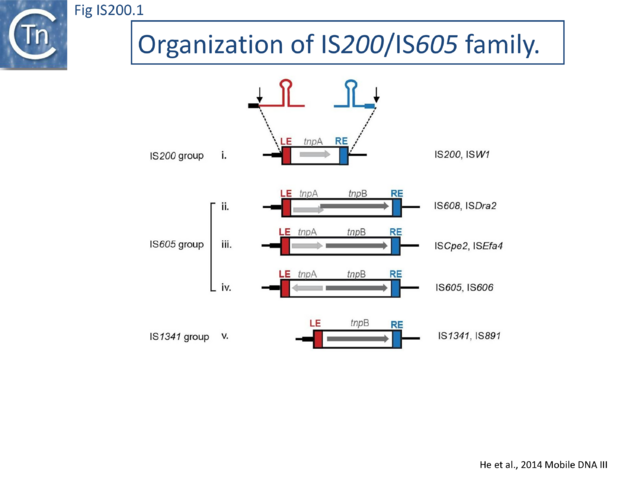
The IS200/IS605 family members transpose using obligatory single strand(ss) DNA intermediates[15] by a mechanism called “peel and paste”. They differ fundamentally in the organization from classical IS. They have sub-terminal palindromic structures rather than terminal IRs (Fig. IS200.1) and insert 3’ to specific AT-rich tetra- or penta-nucleotides without duplicating the target site.
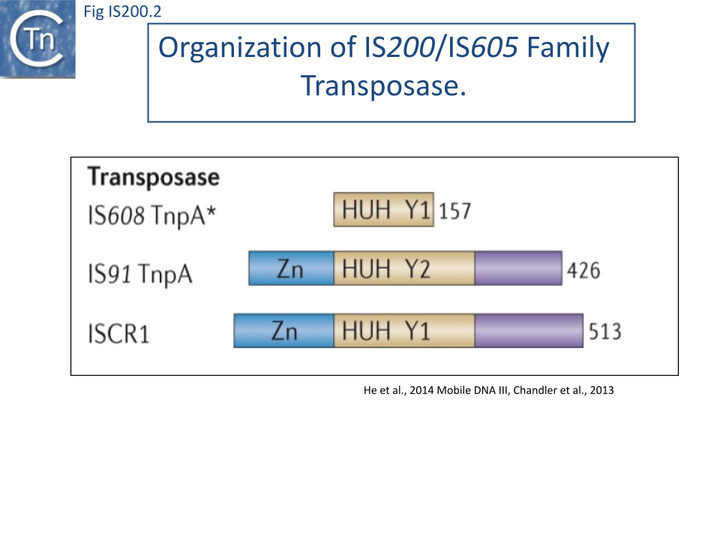
The transposase, TnpA, is a member of the HUH enzyme superfamily (Relaxases, Rep proteins of RCR plasmids/ss phages, bacterial and eukaryotic transposases of IS91/ISCR and Helitrons[16][17])(Fig. IS200.2) which all catalyze cleavage and rejoining of ssDNA substrates.
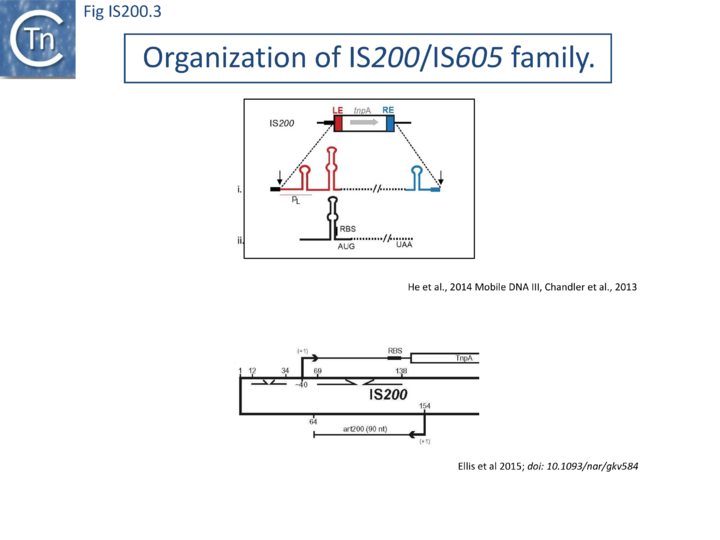
IS200, the founding member (Fig. IS200.3), was identified 30 years ago in Salmonella typhimurium [1] but there has been renewed interest for these elements since the identification of the IS605 group in Helicobacter pylori [11][18][14]. Studies of two elements of this group, IS608 from H. pylori and ISDra2 from the radiation resistant Deinococcus radiodurans, have provided a detailed picture of their mobility [19][20][21][22][23][24][25].
Distribution and Organization
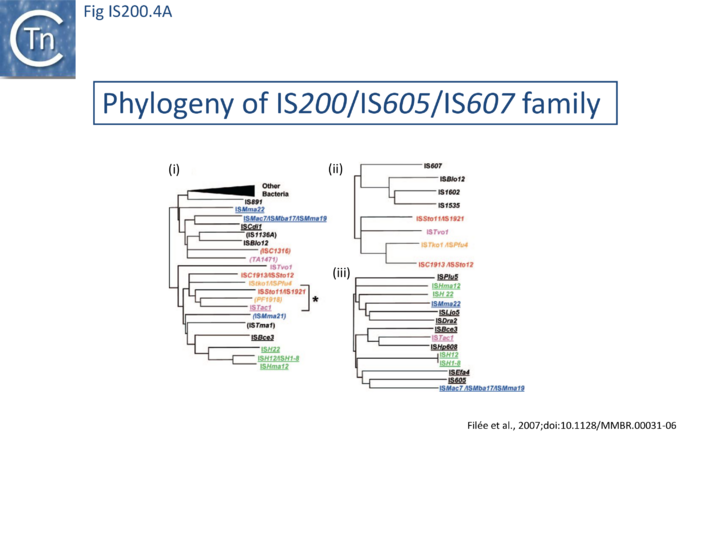
The family is widely distributed in prokaryotes with more than 153 distinct members (89 are distributed over 45 genera and 61 species of bacteria, and 64 are from archaea). It is divided into three major groups based on the presence or absence and on the configuration of two genes: the transposase tnpA (https://www.ncbi.nlm.nih.gov/research/cog/cog/COG1943/), sufficient to promote IS mobility in vivo and in vitro and tnpB (https://www.ncbi.nlm.nih.gov/research/cog/cog/COG0675/) (Fig. IS200.1) initially of unknown function and not required for transposition activity but now known to de an RNA-guide endonuclease (see TnpB below) . These groups are: IS200, IS605 and IS1341. TnpB is also present in another IS family, IS607, which uses a serine-recombinase as a transposase. In the phylogeny of this group (Fig. IS200.4A) of IS, both tnpB and tnpA of bacterial or archaeal origin are intercalated, suggesting some degree of horizontal transfer between these two groups of organisms[26].
Isolated copies of IS200-like tnpA can be identified in both bacteria and archaea[26]. Full length copies of IS605-like elements are also found in bacteria and several archaea and all have corresponding MITEs (Miniature Inverted repeat Transposable Elements) derivatives in their host genomes.
The IS200 group
IS200 group members encode only tnpA, and are present in gram-positive and gram-negative bacteria and certain archaea[2][27] (Fig. IS200.1 and Fig. IS200.3). Alignment of TnpA from various members shows that they are highly conserved but may carry short C-terminal tails of variable length and sequence. Among approximately 400 entries in ISfinder (December 2023), about 50 examples IS200-like derivatives.
They can occur in relatively high copy number (e.g. >50 copies of IS1541 in Yersinia pestis) and are among the smallest known autonomous IS with lengths generally between 600-700 pb. Some members such as ISW1 (from Wolbachia sp.) or ISPrp13 (from Photobacterium profundum) are even shorter.
IS200 was initially identified as an insertion mutation in the Salmonella typhimurium histidine operon [1]. It is abundant in different Salmonella strains and has now also been identified in a variety of other enterobacteria such as Escherichia, Shigella and Yersinia.
Different enterobacterial IS200 copies have almost identical lengths of between 707 and 711bp. Analysis of the ECOR (E. coli) and SARA (Salmonellae) collections showed that the level of sequence divergence between IS200 copies from these hosts is equivalent to that observed for chromosomally encoded genes from the same taxa[28][29]. This suggests that IS200 was present in the common ancestor of E. coli and Salmonellae.
In spite of their abundance, an enigma of IS200 behavior is its poor contribution to spontaneous mutation in its original Salmonella host: only very rare insertion events have been documented [2]. One reason for these rare insertions could be due to poor expression of the TnpAIS200 gene from a weak promoter pL identified at the left IS end (LE)[4][5] (Fig. IS200.3).
Besides the characteristic major subterminal palindromes [4] presumed binding sites of the transposase at both LE and the right end (RE) (Substrate recognition), IS200 carries also a potential supplementary interior stem-loop structure (Fig. IS200.3). These two structures play a role in regulating IS200 gene expression. The first (perfect palindrome at LE; nts 12–34) overlaps the TnpAIS200 promoter pL, can act as a bi-directional transcription terminator upstream of TnpAIS200 and terminates up to 80% of transcripts[30] (Fig. IS200.3). The second (interior stem-loop; nts 69–138) (Fig. IS200.3), at the RNA level, can repress mRNA translation by sequestration of the Ribosome Binding Site (RBS) (Fig. IS200.3). Experimental data suggested that the stem-loop is formed in vivo and its removal by mutagenesis caused up to a 10 fold increase in protein production[30]. Recent deep sequencing analysis revealed another aspect in post-transcriptional regulation of IS200 expression: A small anti-sense RNA (asRNA) IS200 transposase expression (Fig. IS200.3) was identified as a substrate of Hfq, an RNA chaperone involved in post-transcriptional regulation in numerous bacteria[31]. Interestingly, asRNA and Hfq independently inhibit IS200 transposase expression: knock-out of both components resulted in a synergistic increase in transposase expression. Moreover, footprint data showed that Hfq binds directly to the 5’ part of the transposase transcript and blocks access to the RBS[32].
In spite of its very low transposition activity, an increase in IS200 copy number was observed during strain storage in stab cultures[1][3]. However, the factors triggering this activity remain unknown[2] . Transient high transposase expression leading to a burst of transposition was proposed to explain the observed high IS200 (>20) copy number in various hosts and in stab cultures [1].
Although regulatory structures similar to that observed in IS200 (Fig. IS200.3) were predicted in IS1541, another member of this group with 85% identity to IS200, this element can be detected in higher copy number (> 50) in Salmonella and Yersinia genomes. However, no detailed analysis of its transposition is available and since no de novo insertions have been experimentally documented and chromosomal copies appear stable in Y. pestis[33], it remains possible that IS1541 also behaves like IS200.
However, the regulatory structures are not systematically present in other IS200 group members and understanding of the control of transposase synthesis requires further study.
The IS605 group
IS605 group members are generally longer (1.6-1.8 kb) due to the presence of a second orf, tnpB in addition to tnpA. Alignment of TnpA copies from this group indicated that although they do not form a separate clade from the IS200 group TnpA, they generally carry the short C-terminal tail. The tnpA and tnpB orfs exhibit various configurations with respect to each other. They may be divergent (Fig. IS200.1 i top: e.g. IS605, IS606) or expressed in the same direction with tnpA upstream of tnpB. In these latter cases, the orfs may be partially overlapping (Fig. IS200.1 ii; e.g. IS608, ISDra2) or separate Fig. IS200.1 iii; e.g. ISSCpe2, ISEfa4). tnpB is also sometimes associated with another transposase, a member of the S-transposases (e.g. IS607[12][34], see [15]. TnpB was not required for transposition of either IS608 or ISDra2.
Three related IS, IS605, IS606 and IS608 (Fig. IS200.1) have been identified in numerous strains of the gastric pathogen Helicobacter pylori [11][14] . IS605 is involved in genomic rearrangements in various H. pylori isolates[35].
The H. pylori elements transpose in E. coli at detectable frequencies in a standard "mating-out" assay using a derivative of the conjugative F plasmid as a target [11][14].
The two best characterized members of this family are IS608 and the closely related ISDra2 from Deinococcus radiodurans. Both have overlapping tnpA and tnpB genes (Fig. IS200.1 ii). Like other family members, insertion is sequence-specific: IS608 inserts in a specific orientation with its left end 3’ to the tetranucleotide TTAC both in vivo and in vitro[14] while ISDra2 inserts 3’ to the pentanucleotide TTGAT[38]. Interestingly ISDra2 transposition in its highly radiation resistant Deinococcal host is strongly induced by irradiation[39] (Single strand DNA in vivo). Their detailed transposition pathway has been deciphered by a combination of in vivo studies and in vitro biochemical and structural approaches (Mechanism of IS200/IS605 single strand DNA transposition).
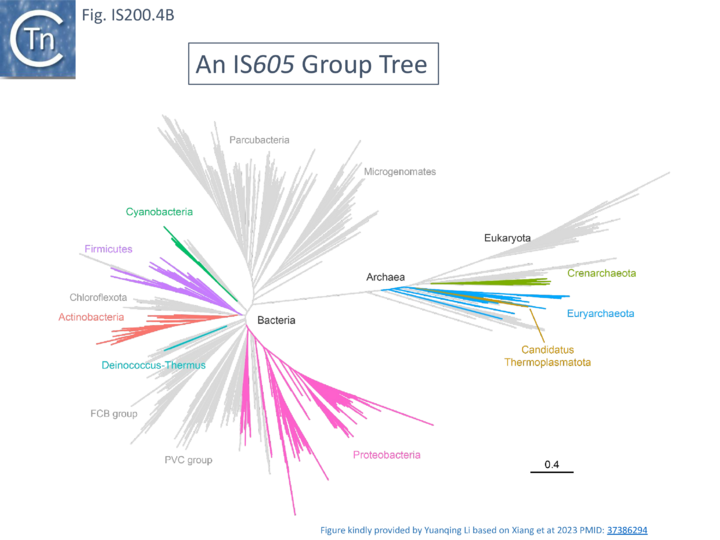
A more detailed and recent analysis of the distribution of 107 IS605 group elements in ISfinder is shown in Fig. IS200.4B [36]. The tree, based on TnpB sequences could be divided into 8 clusters which are overlaid onto the universal tree described by Hug et al., 2016 [37].
The IS1341 group
Elements of the third group, IS1341, are devoid of tnpA and carry only tnpB (Fig. IS200.1 v). The IS occurs in three copies in Thermophilic bacterium PS3 [8]. Multiple presumed full-length elements (including tnpA and tnpB) and closely related copies have been identified in other bacteria such as Geobacillus. On the other hand, IS891 from the cyanobacterium Anabaena is present in multiple copies on the chromosome and is thought to be mobile since a copy was observed to have inserted into a plasmid introduced in the strain[9].
Another isolated tnpB-related gene, gipA, present in the Salmonella Gifsy-1 prophage may be a virulence factor since a gipA null mutation compromised Salmonella survival in a Peyer's patch assay [40]. While no mobility function has been suggested for gipA, it is indeed bordered by structures characteristic of IS200/IS605 family ends and closely related to E. coli ISEc42.
In spite of their presence in multiple copies, it is still unclear whether IS1341 group members are autonomous IS or products of IS605 group degradation and require TnpA supplied from a related IS in the same cell for transposition.
IS decay
Circumstantial evidence based on analysis of the ISfinder database suggests that IS carrying both tnpA and tnpB genes may be unstable. Thus, although members of the IS200 group are often present in high copy number in their host genomes, intact full-length IS605 group members are invariably found in low copy number (P. Siguier, unpublished) (See also TnpB). On the other hand, various truncated IS605 group derivatives appear quite frequently.
These forms seem to result from successive internal deletions and retain intact LE and RE copies. Sometimes, as in the case of ISSoc3, orf inactivation appears to have occurred by successive insertion/deletion of short sequences (indels) generating frameshifts and truncated proteins. For some IS (e.g. ISCco1, ISTel2, ISCysp14, ISSoc3) degradation can be precisely reconstituted and each successive step validated by the presence of several identical copies (P. Siguier, unpublished). This suggests that the degradation process is recent and that these derivatives are likely mobilized by TnpA supplied in trans by autonomous copies in the genome.
Among the approximately 400 IS200/IS605 family entries in ISfinder (December 2023), there are more than 200 examples of IS1341-like derivatives. It was suggested that the IS1341-like derivatives might undergo transposition using a resident tnpA gene to supply a Y1 transposase in trans. There is some circumstantial evidence for transposition of IS1341-like elements. For example, IS891, present in multiple copies in the cyanobacterium Anabaena sp. strain M-131 genome [9] was observed to have inserted into a plasmid which had been introduced into the strain and more recently it has been shown experimentally that IS1341 derivatives can be mobilized by a resident tnpA gene [41] (see The IS1341 Conundrum).
ISC: A group of Elements Related to the IS605 Group
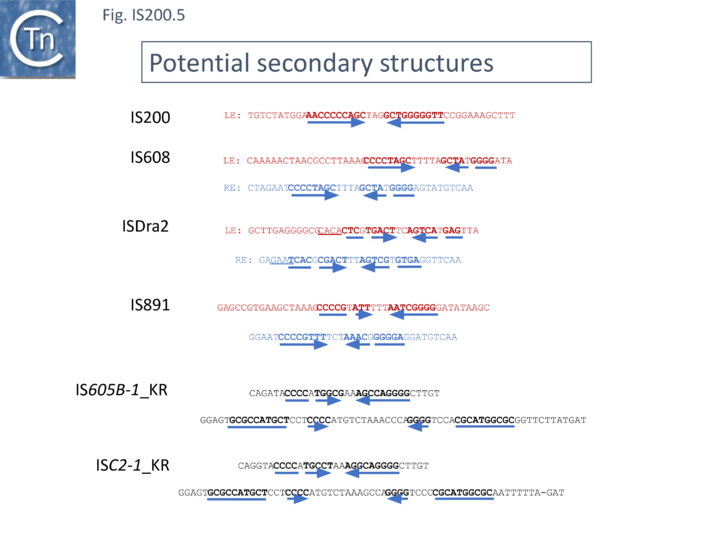
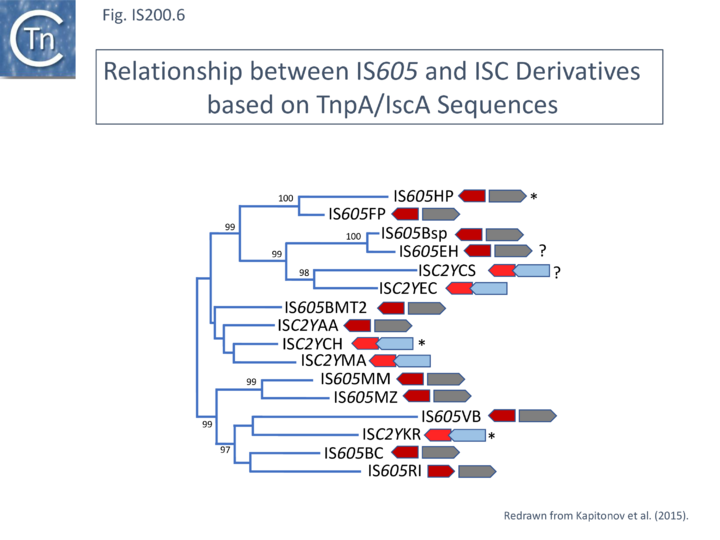
Another group of potential IS of similar organisation, the ISC insertion sequence group, was defined by Kapitonov et al.[42] following identification of Cas9 homologues which occur outside the CRISPR structure, so called “stand-alone” homologues. While related to TnpB, they are more similar to Cas9 than to TnpB proteins. These genes were often flanked by short DNA sequences which, like LE and RE of the IS200/IS605 family, were capable of forming secondary structures. Moreover, it was reported that the ends of many ISC derivatives showed significant identity to members of the IS605 derivatives identified by these authors in the same study. (Fig. IS200.5). These structures therefore resemble the IS1341-like group.
These potential transposable elements were called ISC (Insertion Sequences Encoding Cas9; not to be confused with ISCR, IS with Common Region). The name IscB was coined for the Cas9-like protein and IscA for an associated potential transposase protein which was identified in a very limited number of cases. Examples of ISC elements with both iscA and iscB genes are quite rare. Only 7 cases were identified by Kapitonov et al.,[42] (Fig. IS200.6) and only 56 of 2811 iscB examples observed in a more extensive analysis were accompanied by an iscA copy [43] . Most ISC identified were IS1341-like with only the iscB (tnpB-like) gene. These stand-alone IscB copies were identified in multiple copies in a large number of bacterial and archaeal genomes generally in low numbers (<10 copies) although some genomes contained more elevated numbers (e.g. 22 in Methanosarcina lacustris; 25 in Coleofasciculus chthonoplastes PCC 7420; 52 in Ktedonobacter racemifer)[42].
However, in contrast to the observations of Kapitonov et al.,[42] more wide-ranging studies [43] identified rare IscB proteins which were not “stand alone” but were associated with CRISPR arrays (31 examples in a sample of 2811).
A tree of “full-length” elements (Fig. IS200.6; [42])(i.e. those with both tnpA and tnpB or iscB genes) based on TnpA/IscA sequences showed that full length IS605 and ISC examples carrying both tnpA/iscA and tnpB/iscB are interleaved. IS605 is among those family members with divergent tnpA and tnpB genes (Fig. IS200.1) while other family members carry tnpA upstream of tnpB (e.g. ISDra2). However, in contrast to all IS605-like derivatives, those full length ISC elements included in this tree all have the iscA gene downstream of and slightly overlapping with iscB.
ISC have very similar transposases to those of the IS200/IS605 family and are therefore part of the same super family.
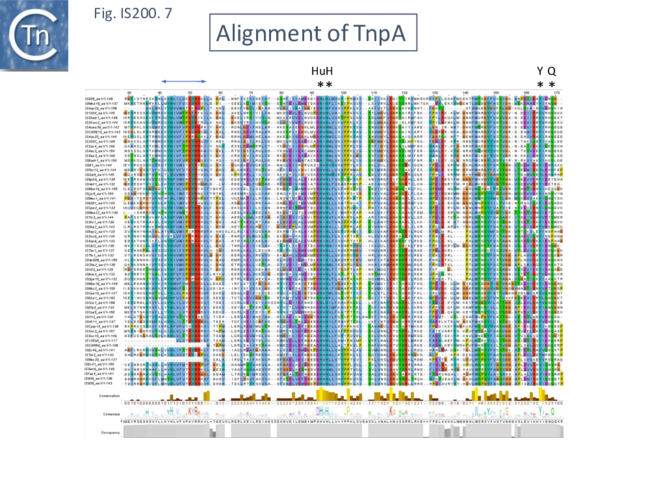
An alignment of full length TnpA from the IS200/IS605 group (Fig. 200.7; ISfinder November 2021) shows the highly conserved HuH triad, catalytic tyrosine (Y) and important glutamine (Q) residues all central to the transposition chemistry (Fig. IS200.7, Fig. IS200.11 and Fig. IS200.12) together with a number of other highly conserved amino acid positions. An alignment with the available IscA from the ISC group (Fig. 200.8 Top) shows that these also include all the highly conserved TnpA amino acid positions and are therefore very closely related to TnpA. However, the IscA and TnpA proteins appear to fall into separate clades (Fig. 200.8 bottom) with some overlap.
Since IS families are defined by their transposases rather than their accessory genes, and those of ISC and the IS200/IS605 family are so similar, it seems reasonable to include the ISC group as a subgroup of the IS200/IS605 family (or IS605 super family;[42] ). For many of the archaeal elements, there is a small, potential 40-45 amino acid, peptide located upstream of the TnpB analogue.
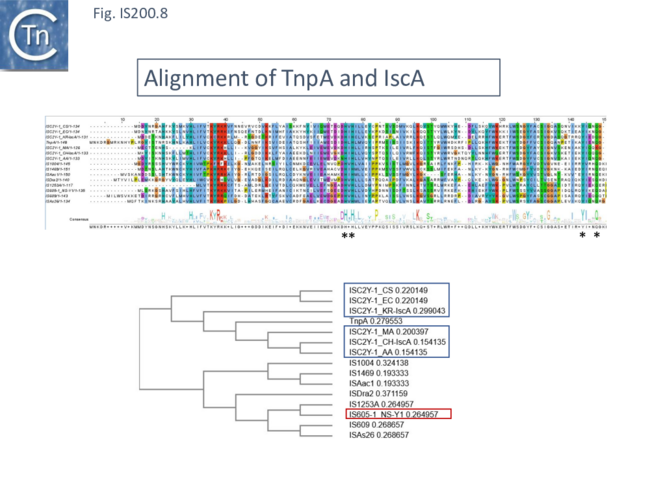
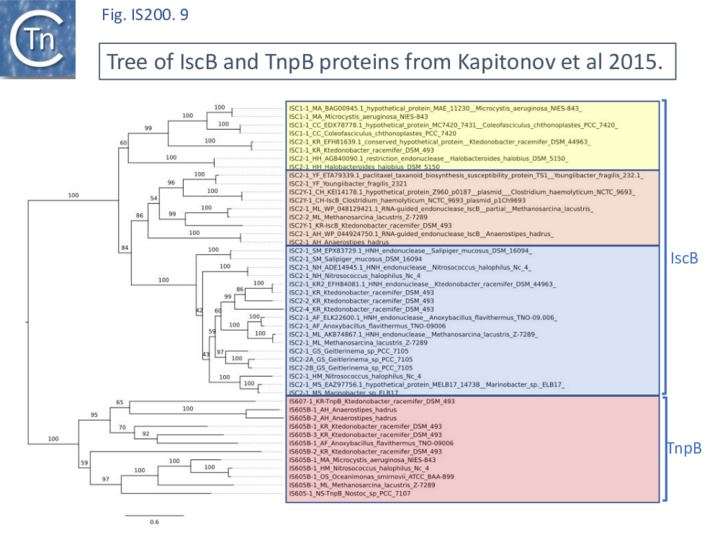
A tree based on the TnpB/IscB (Fig. IS200.9) examples presented by Kapitonov, et al.,[42] shows that the TnpB homologues form a clade separate from IscB and that the latter can be divided into two clades, IscB1 and IscB2.
These considerations therefore reinforce the idea that the IS200/IS605 family and ISC group might be considered as a superfamily which includes a number of related accessory genes (tnpB, iscB1, iscB2 etc), which carry flanking DNA sequences with secondary structure potential and in which a Y1 HuH transposase assures the chemistry of transposition. A similar conclusion was also reached by Altae-Tran et al.[43] .However, this picture is complicated by the identification of another group of transposable elements, the IS607 family in which tnpB is associated with a different type of transposase, in this case a serine site-specific recombinase (IS607 family).
Mechanism of IS200/IS605 single strand DNA transposition
Early models
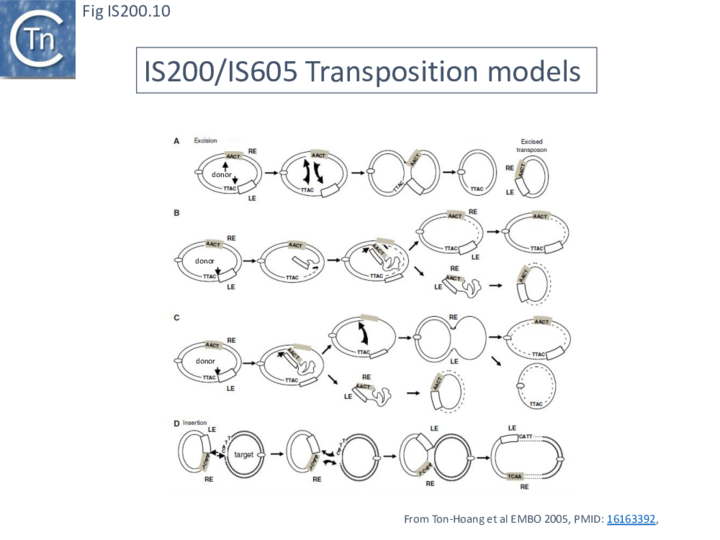
A number of alternative mechanisms were initially proposed to explain IS608 transposition [20] (Fig. IS200.10). These all included the insertion of a double-strand circular transposon copy (Fig. IS200.10 D). One model (Fig. IS200.10 A) envisaged simultaneous or consecutive cleavage at LE and RE and reciprocal strand transfer would generate a Holliday junction (HJ) which then could be resolved into double-strand circular copies of the transposon. The second (Fig. IS200.10 B) cleavage at LE and replicative strand displacement using a 3’OH of the flanking donor DNA. This could assist formation of a single strand region accessible for cleavage of RE to generate a single-strand transposon circle which could be replicated into a double-strand copy. The third (Fig. IS200.10 C) proposed cleavage at LE with displacement of the transposon strand to form a single strand loop. Subsequent in vitro and in vivo experiments (below) demonstrated that not only was IS608 capable of excision as a single-strand DNA circle but that this could be inserted into a single strand target.
General transposition pathway
The transposition pathway of IS200/IS605 family members is shown in Fig. IS200.11. Much of the biochemistry was elucidated using an IS608 cell-free in vitro system which recapitulates each step of the reaction. This requires purified TnpAIS608 protein, single strand IS608 DNA substrates and divalent metal ions such as Mg2+ or Mn2+ [20][21][22]. Similar and complementary results were also obtained with ISDra2[23][24][25]. The reactions are not only strictly dependent on single strand (ss) DNA substrates but are also strand-specific: only the “top” strand (defined as the strand carrying target sequence, TS, 5’ to the IS; Fig. IS200.11 top) is recognized and processed whereas the “bottom” strand is refractory[20] [21]. Cleavage of the top strand at the left and right cleavage sites (TS/CL and CR, note that TS is also the left cleavage site CL) (Fig. IS200.11 B) leads to excision as a circular ssDNA intermediate with abutted left and right ends (transposon joint) (Fig. IS200.11 C bottom left). This is accompanied by rejoining of the DNA originally flanking the excised strand (donor joint).

The transposon joint is then cleaved (Fig. IS200.5 E bottom right) and integrated into a single strand conserved element-specific target sequence (TS) where the left end invariably inserts 3’ to TS (Fig. IS200.5 F). This target specificity is another unusual feature of IS200/IS605 transposition. The target sequence is characteristic of the particular family member and, although it is not part of the IS, it is essential for further transposition because it is also the left end cleavage site CL of the inserted IS [20] (The Single strand Transpososome and Cleavage site recognition) and is therefore intimately involved in the transposition mechanism.
TnpA, Y1 transposases and transposition chemistry
IS200/IS605 family transposases belong to the HUH enzyme superfamily. All contain a conserved amino-acid triad composed of Histidine (H)-bulky hydrophobic residue (U)-Histidine (H)[44] providing two of three ligands required for coordination of a divalent metal ion that localizes and prepares the scissile phosphate for nucleophilic attack. HUH proteins catalyze ssDNA breakage and joining with a unique mechanism. They all catalyse DNA strand cleavage using a transitory covalent 5' phosphotyrosine enzyme-substrate intermediate and release a 3' OH group [17] (Groups with HUH Enzymes; Fig.7.5).
The HUH enzyme family also includes other transposases of the IS91/ISCR and Helitron families as well as proteins involved in DNA transactions essential for plasmid/virus rolling circle replication (Rep; not to be confused with the TnpAREP/REP system described in Domestication) and plasmid conjugation (Mob/relaxase) (Groups with HUH Enzymes; Fig.7.5).
IS200/IS605 transposases are single-domain proteins containing a single catalytic tyrosine residue, called Y1 transposase. They use the tyrosine residue (Y127 for IS608) as a nucleophile to attack the phosphodiester link at the cleavage sites (vertical arrows in Fig. IS200.11 A and D). Since cleavages at both IS ends occur on the same strand, the polarity of the reaction implies that the enzyme forms a covalent 5’-phosphotyrosine bond with the IS at LE producing a 3’-OH on the DNA flank and a 5’-phosphotyrosine bond at the RE flank producing a 3’-OH on RE itself (Fig. IS200.11 B). The released 3′-OH groups then act as nucleophiles to attack the appropriate phospho-tyrosine bond resealing the DNA backbone in one case and generating a single-strand DNA transposon circle in the other (Fig. IS200.11 C). The same polarity is applied to the integration step (Fig. IS200.11 D, E and F). As an important mechanistic consequence of this chemistry, IS200/IS605 transposition occurs without loss or gain of nucleotides. In vitro, the reaction requires only TnpA and does not require host cell factors.
TnpA overall structure
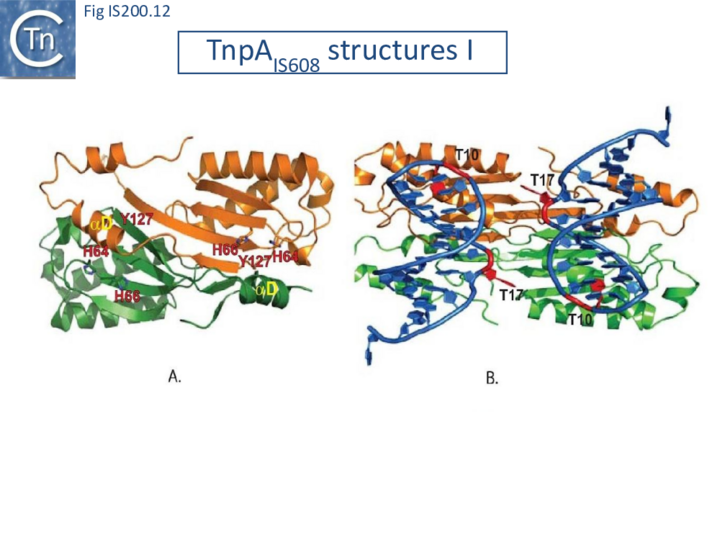
Crystal structures of Y1 transposases have been determined for three family members: IS608 (TnpAIS608) from Helicobacter pylori [19][22] ISDra2 (TnpAISDra2) from Deinococcus radiodurans [25] and ISC1474 from Sulfolobus solfataricus[45]. In contrast to most characterised HUH enzymes, which are usually monomeric and have two catalytic tyrosines, Y1 transposases form obligatory dimers with two active sites (Fig. IS200.12 A). The two monomers dimerize by merging their β-sheets into one large central β-sheet sandwiched between α-helices. Each catalytic site is constituted by the HUH motif from one TnpA monomer (H64 and H66 in the case of TnpAIS608) and a catalytic tyrosine residue (Y127) located in the C-terminal αD helix tail of the other monomer (Fig. IS200.12 A). This is joined to the body of the protein by a flexible loop (trans configuration, Active site assembly and Catalytic activation and Transposition cycle: the trans/cis rotational model).
The TnpA enzyme active sites are believed to adopt two functionally important conformations: the trans configuration described above (Fig. IS200.12 A), in which each active site is composed of the HUH motif supplied by one monomer with the tyrosine residue supplied by the other, and the cis configuration, in which both motifs are contributed by the same monomer (IS200/IS605 video 1 below; kindly supplied by O. Barabas and Fred Dyda).
The trans conformation is active during cleavage where Tyrosine acts as nucleophile whereas the cis conformation is thought to function during strand transfer where the 3’OH is the attacking nucleophile (Transposition cycle: the trans/cis rotational model). Only the trans configuration of TnpAIS608 and TnpAISDra2 has yet been observed crystallographically [19][25] but the existence of the cis configuration is supported by biochemical data [46].
The Single strand Transpososome
The key machinery for transposition is the higher-order protein-DNA complex, the transpososome (or synaptic complex) which contains both transposase and two IS DNA ends with or without target DNA. Transpososome formation, stability, and the temporal changes in a configuration which occur during the transposition cycle have been characterized for TnpAIS608 by crystallographic and biochemical approaches.
Although for technical reasons it was not possible to obtain structures with both LE and RE hairpins together, co-crystal structures with either LE or RE showed that a TnpA dimer binds two subterminal DNA hairpins suggesting that it could bind both LE and RE ends simultaneously. Binding sites for the hairpins are located on the same face of the TnpA dimer while the two catalytic sites are formed on the opposite surface (Fig. IS200.6 A and B) (IS200/IS605 video 2 below; kindly supplied by O. Barabas and Fred Dyda). The hairpin forms a distorted helix anchored by base interactions at the foot (IS200/IS605 video 2 below; kindly supplied by O. Barabas and Fred Dyda).
Substrate recognition
A key feature of TnpA is that it is only active on one strand, the “top” strand. The IS608 and ISDra2 ends carry subterminal imperfect hairpins. In addition to specific sequences on the loops, the irregularities on the hairpins help the enzyme to distinguish between “top” and “bottom” strands [19][25]. The initial co-crystal structure was obtained with TnpAIS608 and a 22nt imperfect RE hairpin (HP22) including its characteristic extrahelical T17 located mid-way along the DNA stem (Fig. IS200.12 and Fig. IS200.13). In addition to a number of backbone contacts with HP22, TnpAIS608 also shows several base-specific contacts, in particular with T10 in the loop and the extrahelical T17[19] (Fig. IS200.12 B).
Exchange of T10 and neighboring T nucleotides in the loop abolished binding whereas the exchange of T17 for an A significantly reduced but did not eliminate binding [47]. Similar studies with TnpAISDra2 showed that it also recognises a similarly located T in the hairpin loop of ISDra2 and that this is essential for binding [25] . Instead of an extrahelical T, ISDra2 LE and RE include a bulge caused by two mismatched nucleotides (G and T) in the hairpin stem. These unpaired nucleotides are specifically recognized and stabilized by the protein. Again, mutation of the T (to C which, in this case, eliminates the bulge to generate a GC base pair in the stem) greatly reduces binding (IS200/IS605 video 3A below; kindly supplied by O.Barabas and Fred Dyda).
Although most members of the IS605 group, which includes IS608 and ISDra2, have imperfect palindromes with extrahelical bases or bulges, some members of the IS200 group (e.g IS200, IS1541) include perfect hairpins. Whether base-specific interactions with the loop sequence is exclusively responsible for strand-specific activity of the corresponding transposase remains to be clarified.
Cleavage site recognition
The left (CL/TS) and right (CR) IS608 cleavage sites (TTACl and TCAAl respectively, where l represents the point of cleavage) are located some distance from the subterminal recognition hairpins (19 nt at LE and 10 nt at RE) (Fig. IS200.13). The system is asymmetric because the two distinct cleavage sites are separated from the hairpins by linkers of different lengths and the CL/TS sequence does not form part of IS while CR does.

Structural studies revealed that the cleavage sites are recognized in a unique way that does not involve direct sequence recognition by TnpA. Instead, an internal part of the IS sequence is co-opted to recognize different cleavage sites allowing TnpA to catalyze both excision and integration of the element with a single DNA binding domain.
Internal transposon sequences, the left (GL) and right (GR) tetranucleotide guide sequences, AAAG and GAAT, located 5’ to the foot of the hairpins (Fig. IS200.7), recognize their respective cleavage sites by direct base interactions. These GL/CL and GR/CR interactions involve 3 of the 4 nt of GL and GR. They include both canonical Watson-Crick interactions and in the case of RE, non-canonical interactions resulting in base triplets (Fig. IS200.13 and Fig. IS200.14, bases joined by both regular and dotted lines respectively). In the case of LE and the transposon joint, base triples (dotted lines) are suggested from biochemical data [47] (IS200/IS605 video 3B below; kindly supplied by O. Barabas and Fred Dyda).
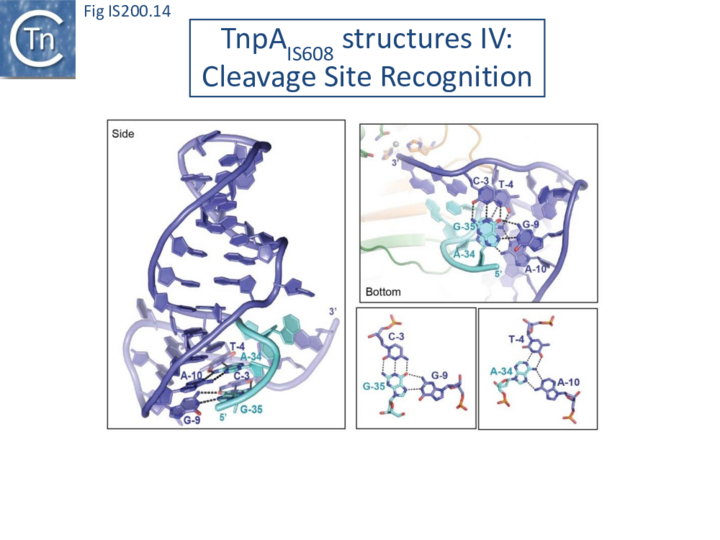
These interactions place the scissile phosphate precisely into the two active sites of TnpAIS608 for nucleophilic attack by the catalytic Y127. Interestingly, the base-pairing patterns responsible for cleavage site recognition are similar at LE, RE and the target site in spite of sequence differences (Fig. IS200.13, Fig. IS200.14, Fig. IS200.15). Since TS is also CL, this type of recognition not only explains the requirement for the TS located at the left end of the inserted IS (Fig. IS200.11, Fig. IS200.15) for further transposition, but also the target specificity. Upon integration, TS is presumably recognized by the GL present on the excised transposon joint. Note that the transposon joint contains only the LE guide sequence GL but not the LE cleavage site CL (Fig. IS200.11, Fig. IS200.15).
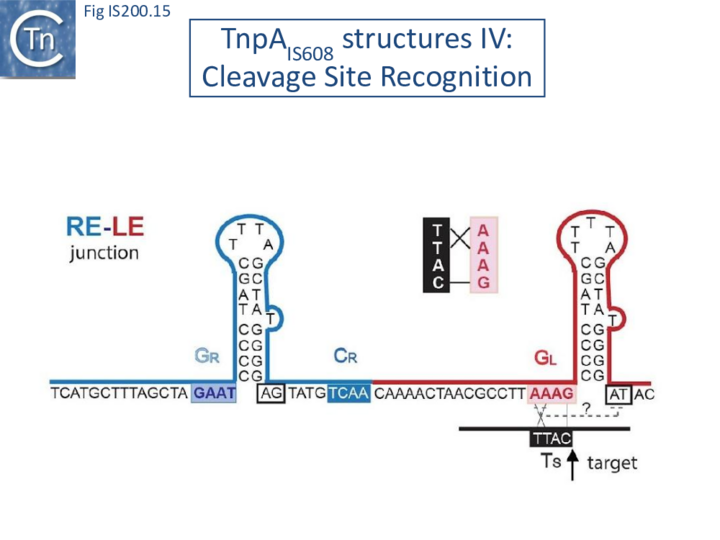
Similar crystal structures were obtained with TnpAISDra2 (see also Single strand DNA in vivo) with a similar interaction network between the guide sequences and cleavage sites.
The ISDra2 transpososome is structurally very similar to those of IS608 despite only 34% sequence identity of the transposases. It is important to note that the target sequence in ISDra2 is a pentanucleotide instead of a tetranucleotide as in IS608. The fifth nucleotide in the ISDra2 sequence is however not involved in DNA-DNA interactions but in DNA-protein interaction[25].
The potential cleavage site recognition mode (i.e. the canonical interaction network between CL,R and GL,R) is indeed well conserved throughout the family (Fig. IS200.16).
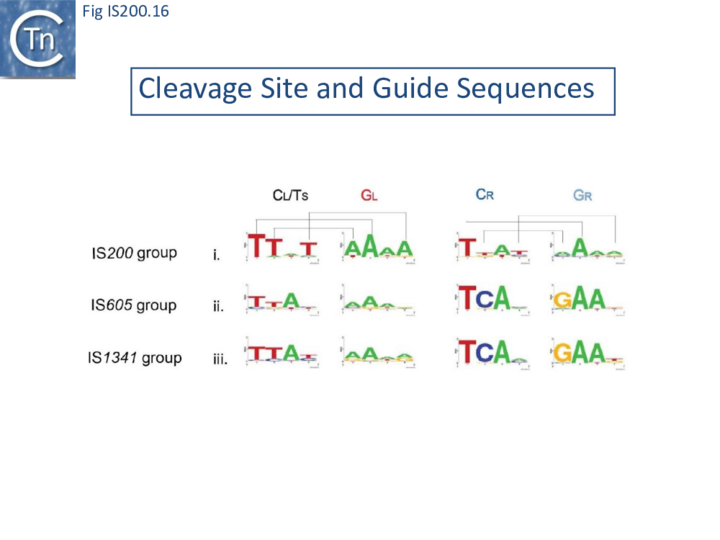
This model has been validated in vitro and in vivo by showing that it is possible to modify cleavage sites by changing corresponding guide sequences. Moreover, in the case of IS608, modifications of GL in the transposon joint generate predictable changes in insertion site-specificity of the element [48]. The IS608 recognition system has also been modified to include additional sequences which assist more specific targeting of insertions[49].
Active site assembly and Catalytic activation
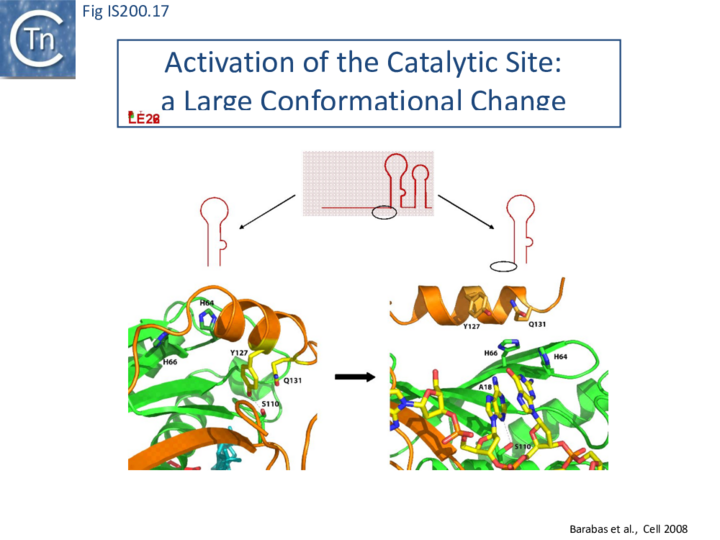
Comparison of crystal structures of different TnpA protein-DNA complexes [19][22] [45] revealed TnpA in both active and inactive configurations. In both the free TnpAIS608 dimer and TnpAIS608-DNA complexes bound to a “minimal” HP22 hairpin (which does not include the guide sequence), the catalytic tyrosine residue (Y127) points away from the HUH motif (H64 and H66) and therefore cannot act as a nucleophile [19] (Fig. IS200.11).
The enzyme is therefore in an inactive conformation. Binding to the appropriate substrate containing the 4 nucleotide guide sequence 5’ to the hairpin foot (compare Fig. IS200.17 left and right) triggers a change in TnpA configuration that permits assembly of functional active sites. A single A (A+18, Fig. IS200.13 and Fig. IS200.13) in the guide sequence present in both GL and GR does not participate in base interactions with the cleavage site. On formation of the CL(R)/GL(R) base interaction network, this single base penetrates the structure and forces the C-terminal αD helix carrying Y127 closer to the HuH motif placing it in the correct position poised for catalysis [22] (compare Fig. IS200.17 left and right; Fig. IS200.18)(IS200/IS605 video 4 below; kindly supplied by O. Barabas and Fred Dyda).
This movement also places a third amino acid (Q131 located at the C-terminal end of helix αD on the same face as Y127) in a position enabling it to function in conjunction with both H residues to complete the metal ion binding pocket. This movement is made possible by the fact that the αD helix is attached to the protein body by a flexible loop. This conformational change involving αD helix movement will be discussed below (Transposition cycle: the trans/cis rotational model).
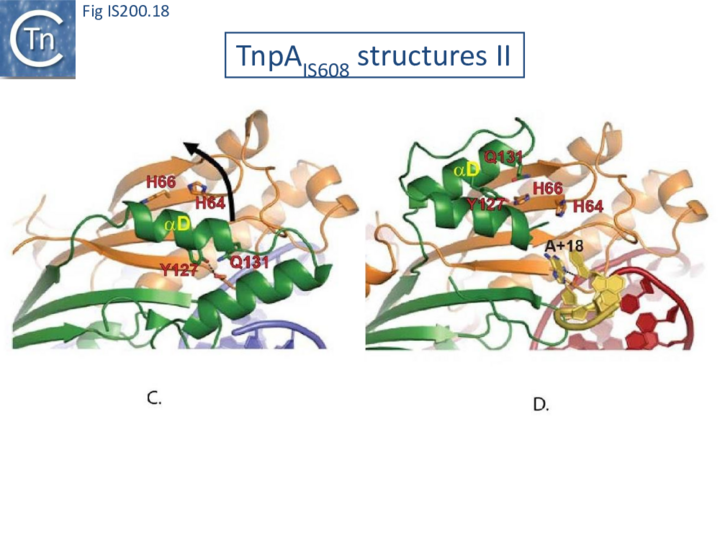
Transpososome assembly and stability
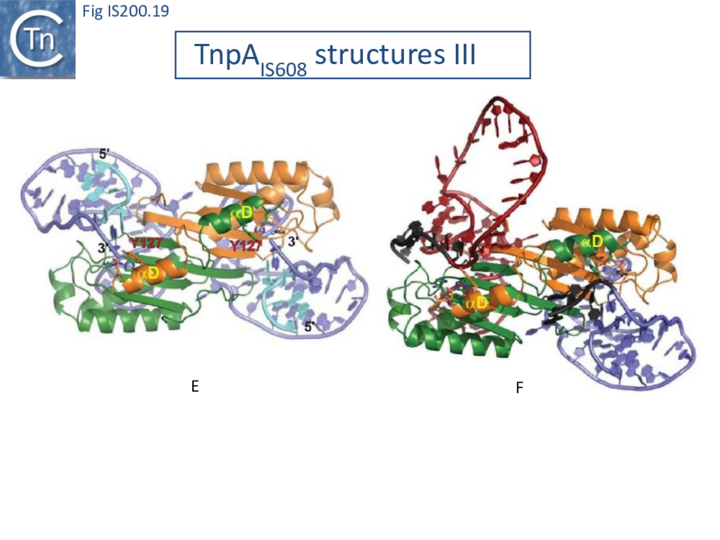
Excision requires the assembly of a transpososome containing both LE and RE. However, it is technically difficult to generate crystallographically pure complexes of this type. Only crystal structures containing two LE or two RE were obtained. The excision transpososome was initially modelled using information obtained from the IS608LE-TnpA and RE-TnpA structures [22] (Fig. IS200.12 B; Fig. IS200.19). However, complexes containing both LE and RE have now been identified using a band shift assay and characterized biochemically [47].
A TnpA co-complex with either LE or RE can be titrated by the addition of increasing quantities of the other end (RE or LE) to obtain a transpososome containing both LE and RE. This can be easily detected in a gel shift assay. Such species proved to be catalytically active since they could be removed from the gel and, when incubated with the essential divalent metal ion, robust reaction products could be detected in a denaturing ge [47].
This approach was used to monitor both transpososome formation and stability using oligonucleotides carrying point mutations in GL,R and CL,R. Robust transpososome formation and cleavage activity requires much of the network of GL,R and CL,R interactions observed in the crystal structures [47] (schematised in Fig. IS200.13). Although base triplets in the original LE co-crystal structure were not detected since the LE substrate was too short [22], the biochemical data suggested that such interactions probably exist (grey dotted lines in Fig. IS200.13).
For example, the two nucleotides 3’ to the foot of the LE hairpin (at equivalent positions to triplet forming bases in RE, Fig. IS200.13 are required for robust synaptic complex formation and cleavage [47]. This further implies that these base triplets might also be involved in target DNA capture (grey dotted lines in Fig. IS200.15).
Base changes in GL resulted in a predictable choice of target sequence [48]. However, large differences in insertion frequencies were observed. The influence of the presumed non-canonical interactions in LE would provide an explanation for this variability since these were not taken into account in the choice of LE guide sequence.
In both IS608 and ISDra2, the extra-helical bases in the hairpin stem and nucleotides in the loop are also important for transpososome formation even in a context which includes both GL,R and CL,R[25][47].
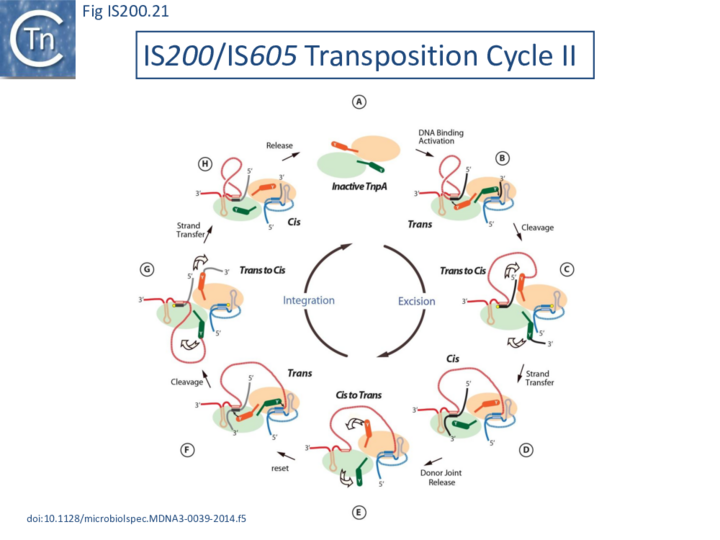
Transposition cycle: the trans/cis rotational model
Transpososome assembly is followed by two critical chemical steps: cleavage and strand transfer. These are thought to be accomplished by a series of large changes in transpososome configuration. A detailed model has been proposed for the dynamics of the IS608 transpososome during the transposition reactions[22] (Fig. IS200.19; IS200/IS605 video 1). As described in TnpA overall structure (above), TnpAIS608 could in principle assume two configurations: trans and cis. Switching between these two states would involve rotation of the two unconstrained flexible arms which join the αD helix to the protein body.
The current model for IS608 and ISDra2 transposition proposes that the strand transfer step involves rotation of these arms from the trans to the cis configuration: cleavage occurs while the enzyme is in the trans configuration. A trans to cis conformational change then occurs allowing strand transfer. The ground state of the IS608 and ISDra2 transpososomes obtained from crystallography is the trans configuration. LE and RE binding and cleavage occur with the enzyme in its trans configuration (Fig. IS200.19; IS200/IS605 video 1).
This results in the formation of the 5’ phosphotyrosine bond with LE liberating a 3’-OH on the flanking DNA and the 5’phosphotyrosine bond with the RE DNA flank liberating a 3’-OH on the RE transposon end. Rotation of the two arms would displace LE towards the sequestered 3’-OH of RE and the RE flank towards the 3’-OH of the LE flank (Fig. IS200.19; IS200/IS605 video 1) and position them so that both 3’-OH can attack the appropriate phosphodiester bond. This model is supported by several lines of indirect evidence from studies of IS608.
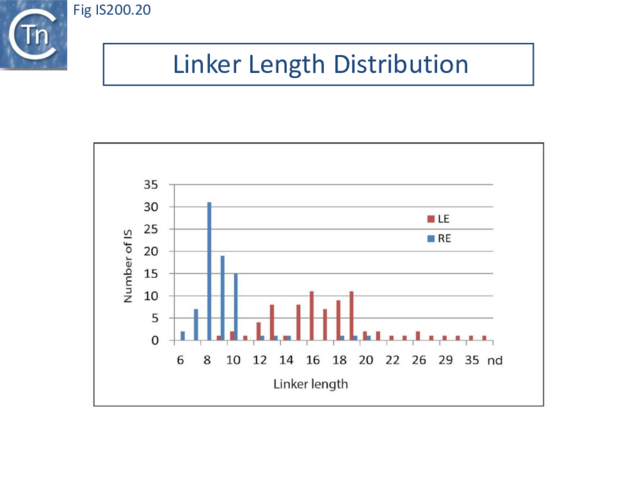
An initial piece of evidence concerns the length differences in the LE and RE “linker” (the distance between the hairpin foot and the cleavage site): this is only 10 nt for RE but 19 nt for LE (Fig. IS200.15). The rotation model suggests that the longer LE linker may be required to provide sufficient length to rotate the 5’ LE phospho-tyrosine bond to position it closes the immobile RE 3’-OH (Fig. IS200.19; IS200/IS605 video 1). This would imply that LE linker length is critical for strand transfer. Indeed, sequential reduction in the length of the LE linker has a large effect on transposition frequency and excision in vivo. In vitro, it also had a somewhat larger effect on strand transfer than on cleavage [47], supporting the idea that the linker is important for mechanical movement.
However, transpososome formation and stability was also observed to be affected with the shortest linkers. This presumably reflects steric barriers to GL(R)/CL(R) interaction and supports the notion that these interactions are important in transpososome assembly. A survey of over 100 different IS from all three groups (35 from the IS200 group; 47 from IS605 and 24 from IS1341) in the public databases has shown that the asymmetry of the IS608 ends is conserved across the entire family: the left linker is always longer than the right (15-16 nt versus 8 nt) [46] (Fig. IS200.20).
The second piece of evidence comes from the behaviour of TnpAIS608 heterodimers carrying point mutations in the HuH or catalytic Y. These were expressed and assembled in vivo and purified based on two different C-terminal affinity tags (one for each monomer). This permitted heterodimers to be distinguished form homodimers. A heterodimer with a combination of mutations that enforce a trans-active TnpA site (in which the wildtype HuH motif and Y127 belong to different TnpA monomers) is proficient for cleavage but not for rejoining. In contrast, a heterodimer with cis-active TnpA site (in which the wildtype HuH motif and Y127 belong to the same TnpA monomer) is proficient for rejoining but inactive in cleavage [46].
This implies that all chemical reactions involved in cleavage occur in the trans site while the chemical reactions for strand transfer occur in the cis site. This strongly supports the rotational model.
A third piece of evidence comes from studies of the flexible arm that joins helix αD to the body of the protein and which is proposed to play a pivotal role in the rotation. This flexibility may be facilitated by two glycine residues (G117 and G118). Mutation of these two residues did not affect strand cleavage but led to inhibition of strand transfer suggesting that the two residues are required for achieving a cis configuration. The importance of these G residues is reflected in their conservation throughout the family [46].
Thus, while the cis configuration has not been observed crystallographically for these elements, its existence is strongly suggested by experimental data, supporting the trans/cis rotational model (Fig. IS200.21).
Regulation of single strand transposition
Single strand DNA in vivo
The obligatory single-stranded nature of IS200/IS605 transposition in vitro suggests that it is limited in vivo by the availability of its ssDNA substrates inside the cells and processes that produce ssDNA may stimulate transposition. We describe below a link between the transposition of these elements and the replication fork. Moreover, in the case of ISDra2, single strand DNA produced during re-assembly of the D. radiodurans genome following irradiation results in stimulation of transposition[23][50]. Transcription or other processes leading to horizontal gene transfer such as transformation, conjugative transfer, or transduction with single strand phages might also favor their mobility.
Replication fork
The replication fork modulates the transposition of many transposable elements (Tn7, IS903, IS10, IS50, Tn4430, P element[51][52][53][54][55][56]. For IS200/IS605 family members, the replication fork, in particular the lagging strand template, is an important source of ss DNA substrates for both excision and integration. Transposition can be considered to follow a “Peel and Paste ” mechanism (Fig. IS200.22) where the IS excises or is “peeled” off as a single strand circle from the lagging strand template of the donor molecule and then integrates or is “pasted” in a ss target at the replication fork.
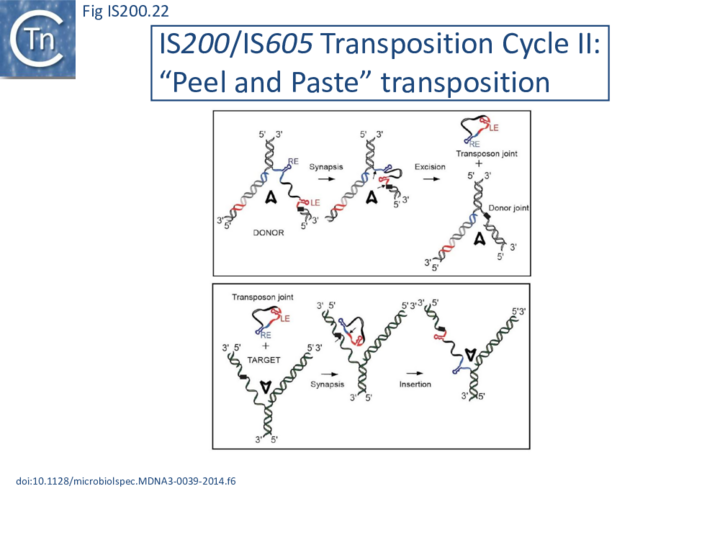
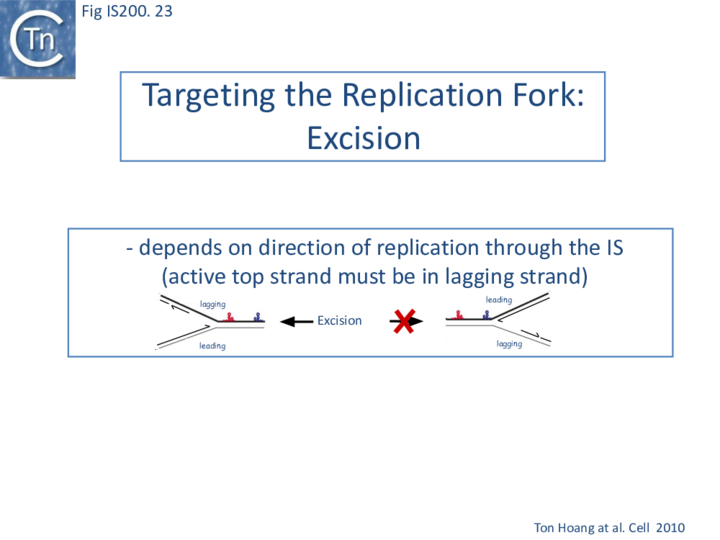
Excision: Excision of IS608 is sensitive to the direction of replication across the element: it is more frequent when the active strand (top strand) is on the lagging strand (discontinuous) template (Fig. IS200.22 top; Fig. IS200.23) but difficult to detect when it is on the leading (continuous) strand [24]. Moreover, excision in vitro requires that both ends are in single strand form at the same time[20].
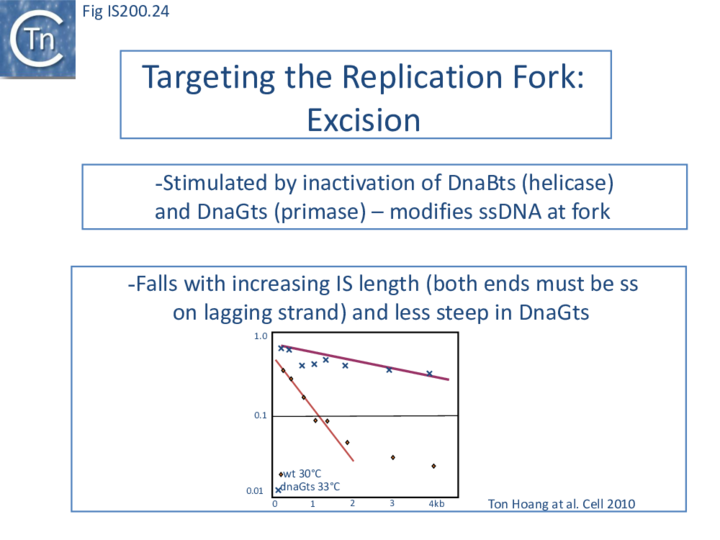
The length of ssDNA on the lagging-strand template depends on the initiation frequency of Okazaki fragment synthesis by the DnaG primase[57][58]. Transient inactivation of DnaG activity reduces this frequency and therefore increases the average length of ssDNA between Okazaki fragments; the IS608 excision frequency increased. Under permissive conditions for E. coli carrying a dnaGts mutation, using a plasmid-based assay with IS608 derivatives of different lengths, the excision frequency decreased strongly as IS length increased. In contrast, when DnaGts activity was reduced by growth under sub-lethal conditions, excision showed a much less pronounced length-dependence (Fig. IS200.24). This length-dependence might also contribute to the difference in copy numbers observed in the IS200 and IS605 groups (see "Distribution and Organization").
Integration: IS608 integration is oriented (with its left end 3’ to a TTAC target site) and it requires an ssDNA target in vitro [14][21]. The close link between transposition and the replication fork is also illustrated by the integration bias, consistent with a preference for an ssDNA target on the lagging strand template (Fig. IS200.22 bottom). This was indeed found to be the case in E. coli for both plasmid and chromosome targets [24]. As expected, the orientation of insertions into the E. coli chromosome was correlated with the direction of replication of each replicore and was consistent with integration into the lagging strand template.
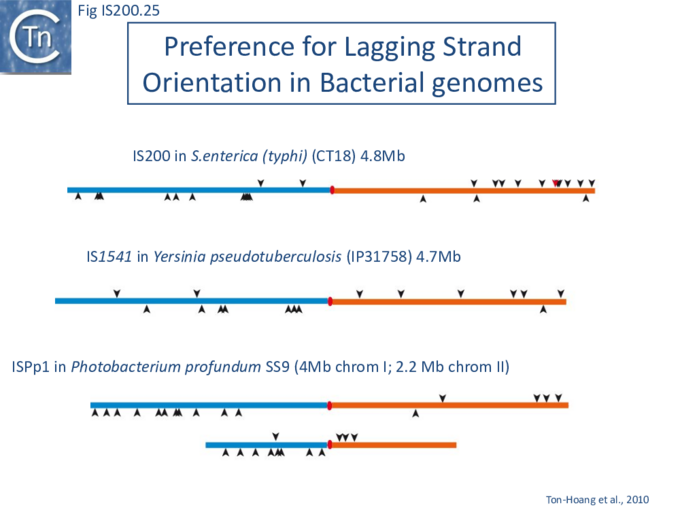
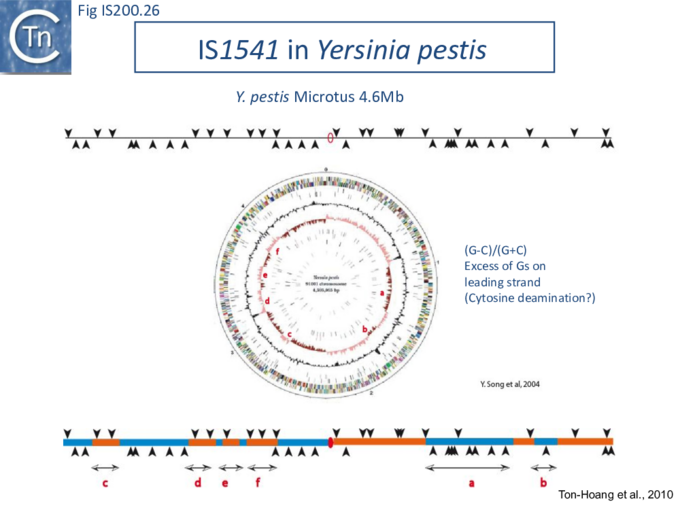
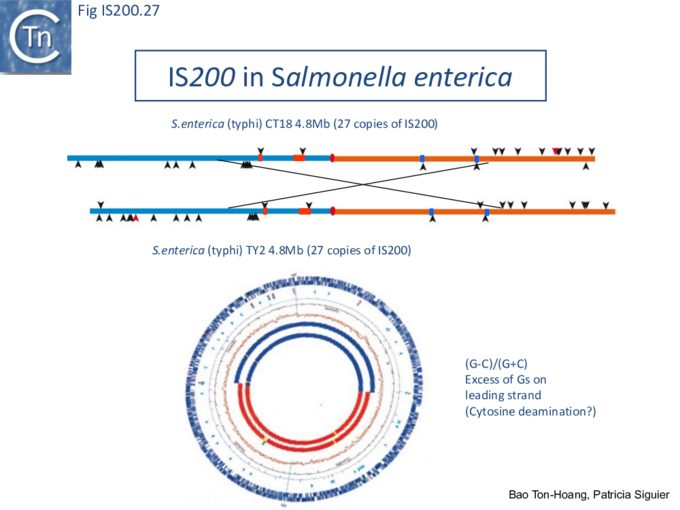
The orientation bias is not restricted to [3]IS608 and ISDra2. An in silico analysis of a large number of bacterial genomes carrying copies of various family members revealed that most had a strong insertional bias consistent with the direction of replication[24] (Fig. IS200.25). Moreover, in certain cases, elements which did not follow the orientation pattern could be correlated to the genomic region that had undergone inversion or displacement (Fig. IS200.26; Fig. IS200.27) suggesting that, once they occur, insertions are quite stable. It seems possible that this type of genomic archaeology based on orientation patterns could be used to complement the study of bacterial genome evolution.
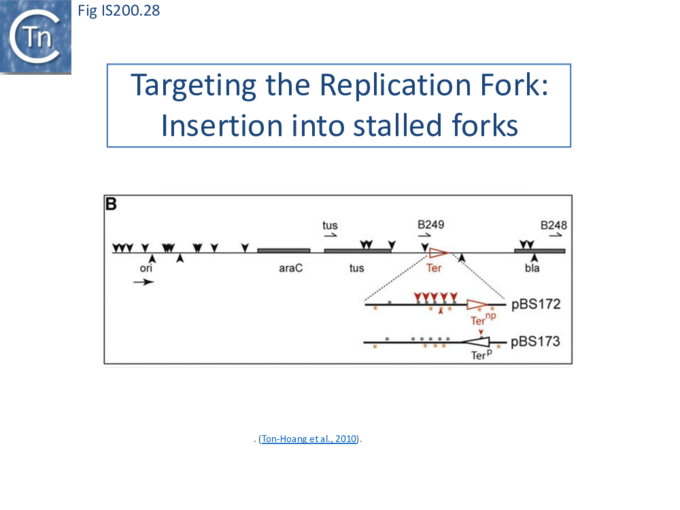
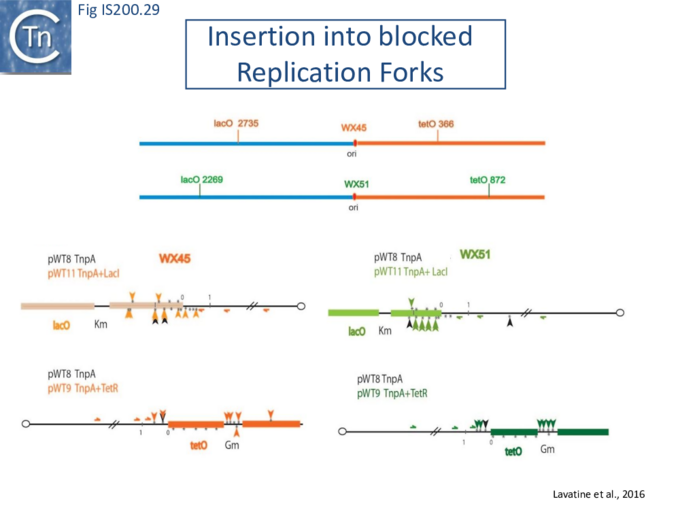
Stalled replication forks: Stalled replication forks appeared preferential targets for IS608 insertion. In the experiments using the Tus/ter replication termination or operator/repressor system, replication fork arrest attracts IS608 insertion [24]. Transient blockade of the unidirectional replication fork by the Tus protein at the ter site resulted in preferential IS608 insertion into the array of target sequences behind the stalled forks on the lagging strand but not on the leading strand (Fig. IS200.28). A similar result was obtained in the E. coli chromosome using the lacI/lacO and tetR/tetO repressor/operator roadblock systems[59][60] (Fig. IS200.29). Moreover, a significant number of IS608 insertions into the E. coli chromosome were localized in the highly transcribed rrn operons. This suggests that high transcription levels might affect replication fork progression (fork arrest by collision with RNA polymerase, R-loop formation, etc.) and could account for targeting the rrn operons. Thus, IS608 insertions can be targeted to the stalled forks and this may well represent a major pathway for targeting transposition.
Genome re-assembly after irradiation in Deinococcus radiodurans
Deinococcus radiodurans, arguably the most radiation-resistant organism known, has a remarkable capacity to survive the lethal effects of DNA-damaging agents, such as ionizing radiation, UV light and desiccation. After exposure to high irradiation doses, the D. radiodurans chromosome which is present in multiple copies per cell[61][62] is shattered and degraded, but can be very rapidly reassembled in a process called ESDSA (Extended Synthesis Dependent Strand Annealing). This involves resection of the multiple dsDNA fragments to generate extensive ssDNA segments, reannealing of complementary DNA and reconstitution of the intact chromosome [39].
Mennecier et al.[50] analyzed the mutational profile in the thyA gene following irradiation. The majority of mutants were due to the insertion of a single IS, ISDra2 which is present in a single copy in the genome of the laboratory D. radiodurans strain. Furthermore, using a tailored genetic system, both ISDra2 excision and insertion efficiency was found to increase significantly following host cell irradiation[23]. A PCR-based approach was used to follow irradiation-induced excision of the single genomic ISDra2 copy and re-closure of flanking sequences. Remarkably, these events are temporally closely correlated with the start of the ESDSA. The signal that triggers ISDra2 transposition is likely the production of ssDNA intermediates generated during genome reassembly. Consistent with this, the requirement of ssDNA substrates for ISDra2, as for IS608, was confirmed by in vitro studies of TnpAISDra2-catalysed cleavage and strand transfer[23].
ISDra2 excision also depends on the direction of replication and is consistent with a requirement for the active strand to be located on the lagging strand template in normally growing cells. However, this bias disappeared in irradiated D. radiodurans [24]. Since no apparent strand bias was observed in generating ssDNA during ESDSA, the lack of orientation bias in irradiated D. radiodurans suggests that ssDNA substrates are no longer limited to those rendered accessible during replication. This indicates that ssDNA sources are different in the contexts of vegetative replication and in genome reassembly.
Real-time transposition (excision) activity
The dynamics of IS608 excision from a donor site has been examined at the colony and single-cell level in real-time using an artificial IS608 derivative inserted between the -35 and -10 elements of a PlacIQ1 promoter[63] driving expression of the blue fluorescent protein mCerulean[64]. TnpAIS608, N-terminally tagged with the bright yellow reporter Venus[65] was supplied in trans driven by PLTetO1 and controllable over a 100x range. Excision rates were proportional to the transposase levels and, as expected, excision depended on the orientation of the IS derivative with respect to the direction of replication in the donor plasmid: IS in an orientation with the active IS strand in the lagging strand template excised more frequently and at lower (10x) TnpA levels than when inserted into the leading strand, demonstrating the validity of the experimental system. In this system, individual excision events as bright flashes of blue fluorescence. Following an initial activity in the part of the population when cells are applied to a solid medium, activity decreases or ceases during “exponential” growth but increases again at a constant rate (in a sub-population) upon growth arrest in a random (Poisson distributed) way. Moreover, the events do not occur randomly in the growing colonies and tend to be excluded from the colony edges. The study underlines the heterogeneity of TE activity rates in both space and time possibly resulting from heterogenous TnpA levels at the individual cell level in the population. These studies are reminiscent of the early studies of Jim Shapiro on phage Mu-mediated rearrangements in growing bacterial colonies[66][67].
TnpB and its Relatives: Guide RNA Endonucleases
TnpA alone can carry out both the cleavage and joining steps in vitro. TnpB is encoded only by the IS1341 and IS605 groups and is not required for transposition of either IS608 or ISDra2 in Escherichia coli and Deinococcus radiodurans respectively [14][20]. The full length TnpB is approximately 400 amino acids long.
IS200/IS605 and the ISC group
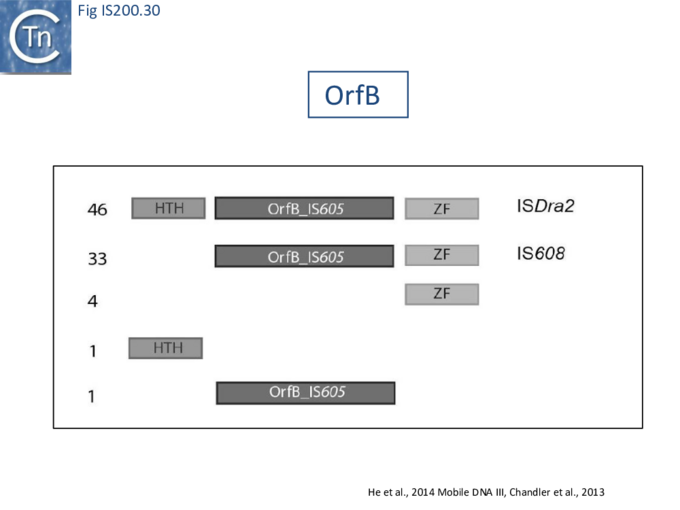
An overview of TnpB organization was originally obtained by comparing the entire ISfinder collection of 85 tnpB copies with the Pfam domain database (Fig. IS200.30). This revealed three major domains: an N-terminal putative helix-turn-helix, a longer and more variable central domain, OrfB_IS605, with a putative DDE motif and a C-terminal zinc finger (ZF) domain of the CPXCG type. Half of the analyzed TnpB copies including TnpBISDra2 but not TnpBIS608 contained all three domains, while only two did not include a zinc finger.
TnpBIS608 was missing the N-terminal HTH domain which would provide an explanation for its lack of activity in certain assays [41].
Pasternak et al.[68] observed that TnpBISDra2 appears to have an inhibitory effect on ISDra2 excision and insertion in its host, D. radiodurans, and on excision in E. coli, and that the integrity of its putative zinc finger motif is required for this effect.
Relatives of TnpB has been identified in both prokaryotes and eukaryotes. It is carried by members of the IS607 family found both in prokaryotes and in eukaryotes and their viruses but is dispensable for IS607 transposition in E. coli . As it is for IS200/IS605 transposition. TnpB analogues, known as Fanzor1 and Fanzor2 (see: Fanzor section below), have also been identified in diverse eukaryotic transposable elements.
TnpB and IscB are Related to the RNA-guided nucleases Cas12 and Cas9.
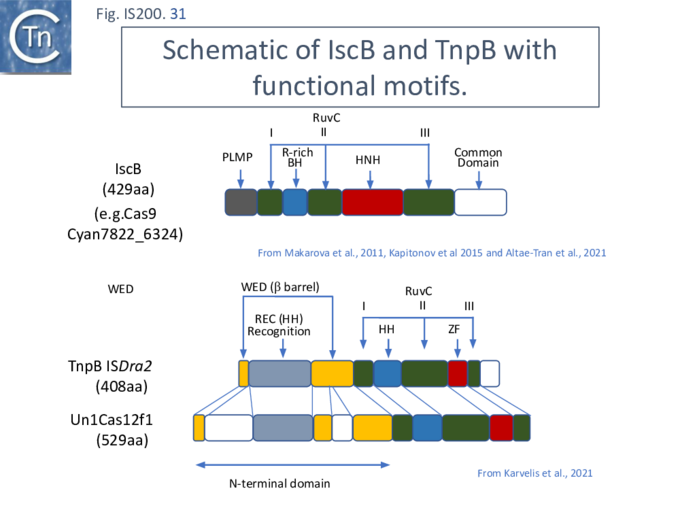
More extensive analysis showed that TnpB shares some similarity with the RNA-guided nuclease Cas12 while IscB showed greater similarity to Cas9. Both, like Cas9 and Cas12, themselves exhibit split RuvC endonuclease domains [42][69] [70][71][72] (Fig. IS200.31). While Cas9 and Cas12 carry related functional domains, their architectures are somewhat different and the configuration of their guide RNAs also differ.
IscB and Cas9
Cas9 (also called Cas5, Csn1, or Csx12) is an RNA-guided dual nuclease generally associated with CRISPR systems in bacteria and widely used in genome engineering. The RuvC DED catalytic triad is split into three sections (I, II and III) in which I and II are interrupted by the R-rich region and II and III by an HNH nuclease domain (Fig. IS200.31). A region common to all Cas9 derivatives is located at the C-terminal end.
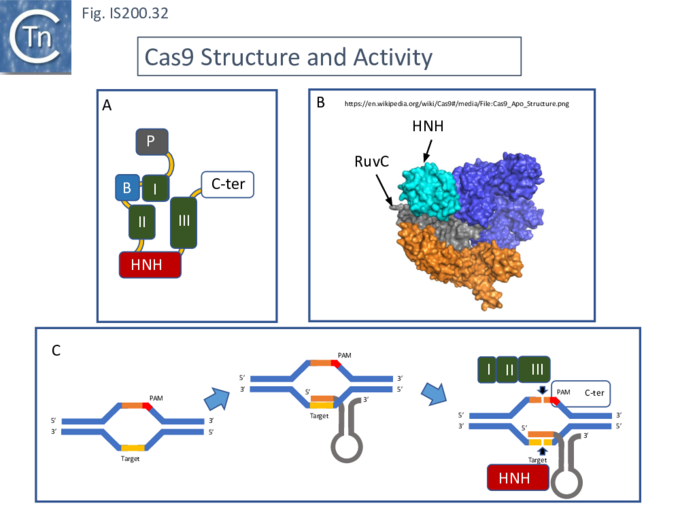
The Cas9 structure has been determined (Fig. IS200.32. B [73]). The protein is a monomer in which the three RuvC segments I, II and II carrying the D, E and D catalytic residues respectively, are assembled into the correct three-dimensional configuration to generate a RuvC-like catalytic pocket with the HNH nuclease domain extruded (Fig. IS200.32. A). The Cas9 guide RNA (crRNA) is composed of a region containing secondary structure potential and a 5’ extension (spacer) of about 20 nts, complementary to the target sequence and which forms an RNA/DNA heteroduplex (Fig. IS200.32. C). Activated Cas9 recognises a specific sequence, PAM (Protospacer Adjacent Motif), located next to the target sequence on the complementary strand downstream of the target sequence. This is necessary for binding of the Cas9-crRNA complex and subsequent cleavage [74]. Cleavage is catalysed by both the HNH nuclease (target strand) and the reconstituted RuvC nuclease (complementary strand). Cleavage is often “blunt” (i.e. occurs at the same position on both strands) and PAM proximal [74].
IscB shares Cas9 sequence features such as the split RuvC and HNH nuclease domains and an arginine-rich (R-rich also known as a bridge helix) domain (Fig. IS200.31 Top) with a group of Cas9 derivatives, Cyan7822_6324, in particular [75]. In addition, a more detailed investigation [43] led to identification of an additional IscB N-terminal domain (called PLMP after its conserved amino acid residues) not present in Cas9 (Fig. IS200.30. Top). These features appear in alignments of IscB sequences [42] ; Fig. IS200.33.

TnpB and Cas12
Cas12 is also an RNA-guided nuclease. A number of subtypes have been described [76] and the structures of several of these have been solved. They have similar C-terminal ends but carry (related) N-terminal ends of various lengths (see Karvelis, et al.[72]). One of the shorter derivatives Cas12F (AKA Cas14) [77] acts as a dimer. Like Cas9, the common C-terminal end is composed of a split RuvC (I, II and III) in which I and II are interrupted by the R/K-rich region. In this case, however, instead of the HNH domain, RuvC segments II and III are separated by a zinc finger of the CPXCG typeI (Fig. IS200.31 bottom).
For Cas12, the guide RNA is composed of a region containing secondary structure potential and a 3’ extension (spacer) of about 20 nts, complementary to the target sequence (Fig. IS200.34). The PAM sequence is located upstream of the target sequence. Cleavage is PAM distal and staggered.
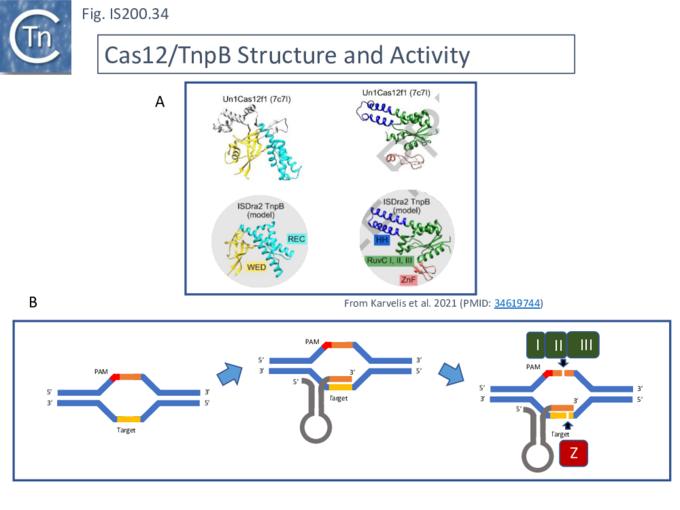
Karvelis, et al.[72] describe the domain structure of TnpB and present evidence that it is related to Cas12, another derivative of the Cas family (Fig. IS200.34 bottom). Like Cas12F, it also carries a RuvC in which the D (I), E (II) and D (III) catalytic residues are split. Again, RuvCI and RuvCII are separated by an R-rich region and RuvCII and RuvCII by a zinc finger with three modules (Fig. IS200.31 bottom). Moreover, the N-terminal region which corresponds to the minimal common structural elements present in Cas12 [72], includes a three helical bundle Rec domain (labelled HTH in an earlier TnpB analysis; Fig. IS200.31 bottom), inserted into a β-barrel domain, referred to as the “Wedge” domain in Cas12. It should be noted that the RuvC domain is used to cleave both DNA strands while the Z domain simply assists this cleavage.
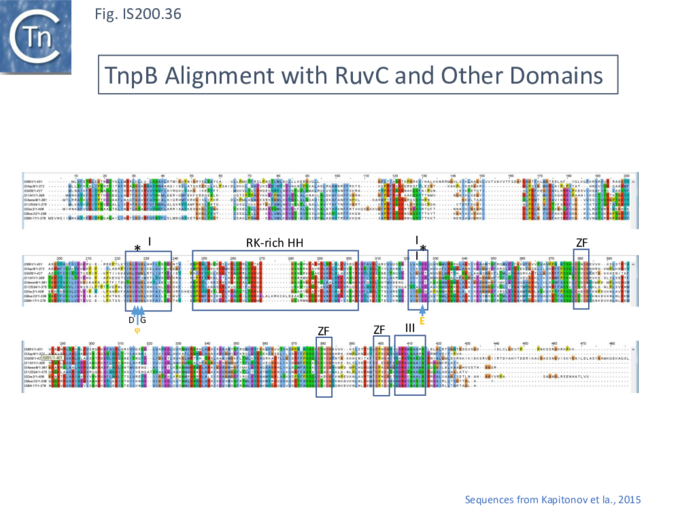
These features can be identified in an alignment of the entire TnpB library (349 examples from ISfinder; November 2021) (Fig. IS200.35 i, ii and iii) and in TnpB sequences provided by Kapitonov et a.,[42] (Fig. IS200.36).
The relationship between Cas12 and TnpB has strong support from structural modelling [72]: for example Un1Cas12f1 (Cas14a) from an uncultured archeon [78], which functions as an asymmetric dimer and represents a minimal domain organization of the Cas12 group [72]. However, TnpB from ISDra2 (see below) appears to be a monomer [72].
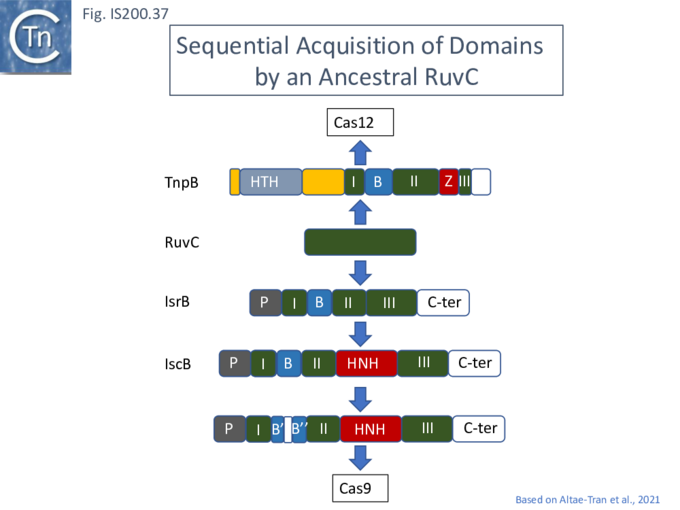
Evolution of TnpB and IscB from an Ancestral RuvC?
In view of the relationship between TnpB, IscB, RuvC and the Cas proteins, the important question of the evolutionary trajectory of these proteins arises. Using various analytic tools, it was concluded that all Cas9 examples identified to date are probably descended from a single IscB derivative ancestor [43]. This contention arose from the observation that the CRISPR-associated IscB derivatives do not form a single clade but are distributed over the IscB phylogenetic tree suggesting that they evolved independently from a single acquisition [43]. Additional IscB derivatives were also identified in this study which led to an evolutionary scenario involving successive acquisition of domains by an ancestral RuvC (Fig. IS200.37). The additional species included a shorter derivative, IsrB, which carried the bridging helix but not the HNH domain and a longer derivative which had acquired a so-called REC domain [43].
TnpB appears to have followed an alternative evolutionary route towards Cas12. In addition, it is thought that TnpB was an ancestor of the eukaryotic Fanzor proteins [80](see: Fanzor section below) associated with diverse eukaryotic potential transposable elements.
Functional analysis of TnpB and IscB
Clearly, the relationship between TnpB and IscB and Cas12 and Cas9 respectively suggested that TnpB and IscB might function as RNA guided nucleases which may, in some way, be involved in transposition [43][72] and this has been extensively tested.
TnpB functions as an RNA-guided Endonuclease
For TnpB, Karvelis, et al.[72] used ISDra2 as a model system. This has the advantage that its transposition behavior has been well characterized [23][68].
In ISDra2, the 3’ end of the upstream tnpA gene overlaps the 5’ end of tnpB. The authors were unable to efficiently express TnpB as a fusion protein but observed that its yield was significantly increased when in its natural context but in which TnpA had been inactivated by mutation. Although the nature of the mutation is not specified in the article, its behavior could be explained if it were an in-frame deletion or other mutation which does not affect C-terminal translation since it seems likely that expression of TnpB involves translational coupling [81][82] with TnpA suggested by their overlapping reading frames (Fig. IS200.38).

TnpB was found to purify with RNA of approximately 150 nts derived from the IS RE (reRNA). reRNA was complementary to the tnpB 3’ end, RE and about 16 nt of (host) flanking DNA (Fig. IS200.38). This RNA with the secondary structure provided by the RE sequence and the 3’ extended flanking DNA is of the expected configuration for relatives of Cas12 (Fig. IS200.36). Previous studies had identified non coding RNA (ncRNA) from the 3’ end of IS1341, a related IS from Halobacterium salinarum NRC-1, called sense overlapping transcripts (sotRNAs) [83].
ncRNAs, sotRNAs and reRNAs
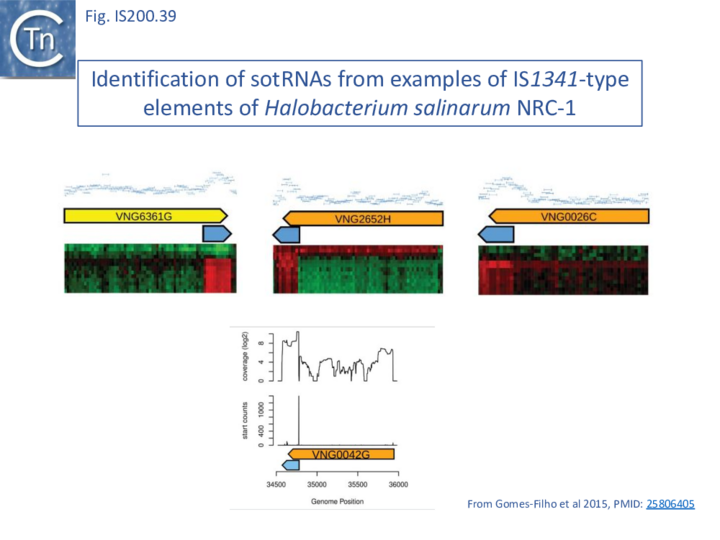
There has been much interest in non-coding RNA (ncRNA) and global searches in Archaea had revealed ncRNA expressed from IS1341 group members which carry only a tnpB gene and are devoid of the TnpA transposase [84][85][86].
During a detailed analysis of ncRNA produced from Halobacterium salinarum NRC-1 [87][88], an ncRNA from the region encompassing the right end of these IS200/IS605 family members was identified. This was called sotRNA (sense overlapping transcript). The authors demonstrated from a publicly available transcriptome compendium [89] that all 10 IS1341 group members in H. salinarum express a sotRNA (Fig. IS200.39) and show condition-dependent differential regulation between sotRNAs and their cognate genes. sotRNA started within tnpB at approximately 1100 nt from its initiation codon, had an average size of 218 nt, and ended approximately 74 nt 3’ to the tnpB termination codon. The authors could not distinguish between the hypotheses that sotRNAs are generated by primary transcription or by processing (although they were unable to locate any potential promoter).
Such sotRNA transcripts, specific for tnpB genes, had previously been identified by Gomes-Filho et al., [87] in a number of Archaea and Bacteria including S. acidocaldarius, Methanopyrus kandleri, Helicobacter pylori and E. coli K12. There has also been some indication of “transposase-related” sense overlapping transcripts of tnpB-like genes from T. kodakarensis [90] and P. furiosus, [91]. However, that these may represent guide RNAs had not been explicitly considered.
Furthermore, sotRNA included what the authors called an RE-like tetraloop resembling the RE DNA loop structure as do sotRNA from P. abyssi and other thermococcal genomes [85].
TnpB: mechanism of action
Karvelis et al.[72] demonstrated that TnpB, purified using a His tag, could cleave DNA. They argued that since the 3’ end of the ISDra2 reRNA corresponds to the DNA target, it would vary according to the position of the IS insertion and the reRNA may (have) serve(d) as a guide RNA. If true, cleavage of the target DNA should occur within the 3’ extension sequence of the flank (the foot of RE in Fig. IS200.38). In this context, it is interesting that the (DNA) structure of the right end was shown to form a base triple which is a characteristic of RNA [21].
To determine whether RNA-guided cleavage occurred , they constructed a system (Fig. IS200.40) using a plasmid supplying TnpB together with an reRNA (Fig. IS200.40 A) which included a 16 (or 20) defined flank sequence and was terminated by a specific Hepatitis delta virus ribozyme (HDV; [92]) to produce a defined 3’ RNA end [93]. A lysate from the host strain was then used in cleavage assays of a library of target plasmids each containing a specific defined 16 base pair sequence directly downstream from a 7 bp (7N) randomised sequence (Fig. IS200.40 B). This has previously been used to identify conserved PAM sequences [72] [94]. Specific double strand cleavage products were captured by adapter ligation (details in Karvelis et al.[72] and the sequence of the resulting enriched 7N region was determined. This corresponded to the conserved ISDra2 target pentanucleotide TTGAT (with a higher enrichment for GA) sequence which is essential for IS insertion and abuts LE in the integrated IS. By equivalence to PAM, this sequence was called TAM (Transposon Adjacent Motif) [72] see also [43] (Fig. IS200.40 C).
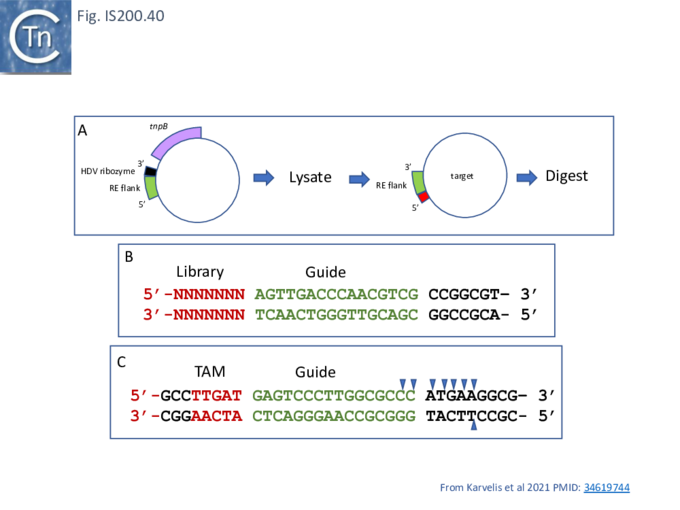
This cleavage specificity was confirmed using purified TnpB-RNP in which the protein and RNA components were produced by separate plasmids and a target plasmid carrying a 3’ flank and a 5’ TTGAT TAM pentanucleotide and a different guide sequence (Fig. IS200.40 C). The results showed a majority of double strand breaks in the supercoiled target plasmid to generate linear plasmids but also a significant level of nicked product. The TnpB-RNP was also active on a linear substrate. In both cases, use of a TnpB D191A mutant, part of the conserved RuvC DED catalytic triad, eliminated the reaction. Robust TnpB-mediated cleavage activity was observed and required both TAM and guide RNA sequences. Further sequence analysis revealed that cleavage occurred distal to the TAM sequence at the guide sequence boundary and was specific for cleavage on the bottom strand but showed some variation on the top strand (Fig. IS200.40). There are some differences however with Cas12. TnpB is a monomer and requires a single copy of reRNA [72].
A similar study by Altae-Tran et al.[43] using purified TnpB from a less well characterised tnpB gene of Alicyclobacillus macrosporangiidus, (AmaTnpB), showed that the protein catalysed cleavage of both double- and single-stranded DNA targets in both a TAM-dependent and TAM independent manner. As in the case of TnpBISDra2, A. macrosporangiidus TnpB-associated guide RNA was identified and derived from the 3’ end of the tnpB gene. In this case, the TAM appeared to be the tetranucleotide TCAC.
These studies therefore identify CL (which is outside the transposon but necessary for transposition by interacting with GL Fig. IS200.13) as the TAM.
An explanation of the “inhibitory effect reported for TnpB?
Moreover, in vivo, TnpB expression together with reRNA from one plasmid resulted in loss of a second plasmid carrying the reDNA target (interference), presumably as a result of cleavage at the target site and linearization of the plasmid. This of course may explain the inhibitory effect of TnpB originally observed by Pasternak et al. [68].
A system which functions in Eukaryotes
Additionally, the authors were able to demonstrate that the system functions in eukaryotic cells opening the possibility that it could be suitably modified for gene editing.
RNA Nomenclature, Processing, Structure, Diversity and mode of function
IS605 group guide RNAs have been called both reRNA and ωRNA (OMEGA for obligate mobile element-guided activity). Here, to eliminate confusion, we will use the term re(ω)RNA (or ω (re)RNA) for that from both groups although they have different secondary structures and functions.
Generating re(ω)RNA: Processing
The important question of how re(ω)RNA is generated was addressed by Nety et al. [95]. Given that TnpB is thought to be an ancestor of Cas12 [96][97], the ability of Cas12 to process RNA (e.g. [96]) may have originated from analogous functions in TnpB [95]. They demonstrated that a TnpB orthologue from the bacterium, A. macrosporangiidus (AmaTnpB), has RNA processing activity and can generate an re(ω)RNA.
The purified AmaTnpB (either wildtype or a RuvC-II catalytic mutant) was incubated with four different in vitro transcribed RNA substrates (Fig. IS200.41 i and ii) produced from PCR-generated DNA templates: a “random” negative control of 1190 nt (Fig. IS200.41 i1); a 166 nt RNA with the RNA guide very similar to that found to be associated with an AmaTnpB orthologue, a potential re(ω)RNA (Fig. IS200.41 i2); a full length tnpB transcript extended to include the guide sequence of 1190 nt (Fig. IS200.41 i3); and the potential re(ω)RNA with a 59 nt 3’ extension of 225 nt (Fig. IS200.41 i4).
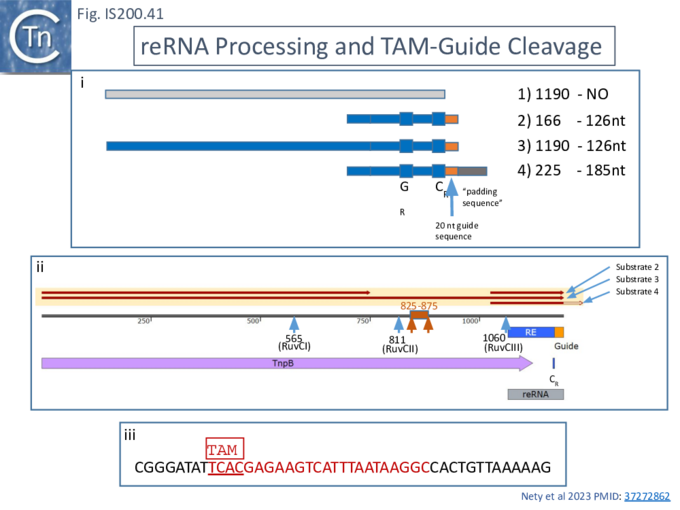
While substrate 1 was refractory to processing, both substrates 2 and 3 generated a 126 nt fragment. Substrate 4 generated a 185 nt fragment suggesting that, while it was processed correctly at the 5’ end, the 3’ extension was not processed. These conclusions were confirmed by RNAseq. All substrates were refractory to the AmaTnpB RuvC-II mutant.
DNA cleavage activities were assessed by including a 1221 nt dsDNA substrate containing the AmaTnpB TAM (Fig. IS200.41 i). RNA substrates 2, 3 and 4 all catalyzed TnpB-mediated DNA cleavage. These results are consistent with those obtained with Dra2TnpB (see below;[98][99]) showing that only the proximal 12 nt of the guide sequence is sufficient for DNA targeting.
The cleavage activity of the three substrates was not identical. The activity of substrate 3, which carries a substantial 5’ extension, was significantly lower than the other two raising the question of whether the extension may include inhibitory sequences.
To investigate this, RNA samples were prepared with different 3’ deletions (Fig. IS200.41 ii) When these RNA species were included in the cleavage reactions, a region between co-ordinates 825 and 875 which shows extensive complementarity to the re(ω)RNA scaffold was observed to be responsible for the inhibitory effect.
This suggests a cis-regulatory mechanism engaged in controlling re(ω)RNA activity [95].
Using ISDra2 [23], Nakagawa et al.,[98] observed that, although TnpB was co-expressed with a 247 nt re(ω)RNA in their purification system, it remained bound to only 100-160 nt of the RNA even in a denaturing gel. Further analysis revealed that the RNA was rapidly degraded in the absence of TnpBDra2 but, in its presence, three different RNAs of approximately 220, 160 and 130 nt were observed, the latter two included the guide sequence at the 3’ end. Very little of the 200nt species was observed in the purified RNP, suggesting degradation, but LC–MS analyses suggested that the 160nt species was cleaved between co-ordinates −150 and −149 or −138 and −137 by TnpB and/or endogenous RNases. They also provide evidence that the ~130-nt RNA is cleaved between −117U and −116G (Fig. IS200.41 ii).
Furthermore, Sasnauskas et al., [99], observed that an re(ω)RNA from between co-ordinates -130 and + 16 was active in DNA cleavage. Nakagawa et al.,[98] also found that truncation of the 5′ region of the re(ω)RNA (−231G to −117U) had no effect on TnpB-mediated DNA cleavage.
Thus re(ω)RNA of ISDra2 also appears to be processed at its 5′ end, and at least a 130 nt fragment including the 3’ guide are stably bound to the TnpB protein.
Structure of TnpB-reRNA in association with DNA
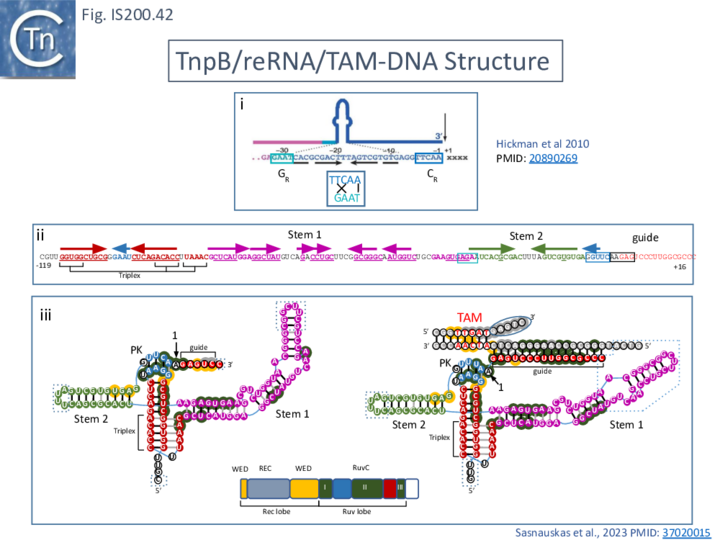
Two studies addressed how TnpB interacts with its DNA template [98] [99] both used TnpBDra2. (Fig. IS200.42). and an re(ω)RNA which included nucleotides -130 to + 16 of the right end (Fig. IS200.42 ii) [99]. Nakagawa et al., [98] used a substrate which was slightly extended in the 5' direction. Both sets of results were essentially the same.
The RNP structure and the ternary structure with the target sequence TnpB could be divided into two “lobes” [98][99]: an N-Terminal lobe (Recognition or Rec) comprising the wedge (WED) and REC domains and a nuclease lobe (Nuc) (insert in Fig. IS200.42 iii) in which the three individual RuvC domains adopt an RNase H fold including D191 (RuvC I), E278 (RuvC II) and D361 (RuvC III).
The results showed that in the RNP complex (Fig. IS200.42 iii left), the principal interactions are with the RuvC and WED domains whereas in the ternary structure with target DNA (Fig. IS200.42 iii right), not only does WED interact with TAM but the RecA domain intervenes around the branch point and the RuvC domain interacts extensively with the target-guide RNA hybrid helix. Note that the CR (TAM) sequence which interacts with GR as DNA during TnpA-mediated transposition ( Fig. IS200.42 i) also forms a short interaction with a sequence upstream which is identical to GR (Fig. IS200.42 ii) to generate a pseudoknot. The scaffold core is formed by the RNA triplex region delimited by the pseudoknot while stem 1 and stem 2 protrude in opposite directions (Fig. IS200.42 iii).
All five TAM positions (Fig. IS200.42 iii right) are recognized directly by the WED domain and substitutions at any TAM position eliminates both target DNA binding and cleavage [99].
On the other hand, substitutions in the guide sequence do not prevent TnpB binding but prevent cleavage. The re(ω)RNA–target DNA heteroduplex (Fig. IS200.42 iii right) is accommodated within a central channel formed by the WED, REC and RuvC domains [98][99].
The authors conclude from the structural results that, for cleavage, the system senses formation of a (perfect) B-form RNA-DNA hybrid without any mismatches because of the effect of guide substitutions and that TnpB requires a 12–16-bp long target perfect DNA-guide RNA heteroduplex to initiate DNA cleavage.
Additional information concerning activity was provided in a study principally exploring diversity in this system (Exploring and defining TAM sequences in the 64 TnpB ISfinder IS605 Group Members).
Xlang et al [36] analyzed re(ω)RNA activity requirements of ISDra2 and three additional IS: ISTfu1, ISDge10 and ISAba30. In these experiments, the 3’ re(ω)RNA scaffold end was defined as the RE tip (Fig. IS200.44).
Activity was exquisitely sensitive to the integrity of CR. Deletion or mutation of all but the 3’ terminal CR base pair significantly reduced activity.
Additionally, the length of the guide sequence was important as was its sequence matches with the target. Optimal editing efficiency occurred with guide sequences between 16 and 20 nucleotides and subsequently decreased with increasing length but was observed to vary somewhat between the three IS (Fig. IS200.42 ii).
Similarly, introduction of single and double base pair transversions into the target, especially in the TAM proximal region approximately up to base pair 12, severely reduced or eliminated activity (Fig. IS200.42 ii) with some variation between the different IS.
This is similar to results obtained with Cas9 and Cas12 systems themselves [100][101]. Finally, variation in 5’ length showed that shortest active scaffolds were 120–140 nt long and lengths of 300 nts were active.
For Dra2TnpB, the C-terminal domain (residues 376 to 408; Fig. IS200.42 bottom insert) has relatively low sequence similarity among TnpB proteins and is disordered in the structures. The C-terminal truncation mutant (Δ376 to 408; ΔCTD) is efficient in target DNA cleavage but exhibits somewhat reduced protein stability. Thus the CTD is not required for RNA-guided target DNA cleavage.
TnpB-re(ω)RNA: Diversity and Activity
In view of the minimal size of the TnpB family guide endonucleases, they may prove useful for targeting applied for biotechnological purposes. It is therefore of importance to determine the extent of their diversity and inherent activities. It had been reported that the TnpB family is an order of magnitude more diverse than the IscB family and an HMMER search of prokaryotic genomes identified >106 tnpB loci [43].
At least two studies [36][95] have addressed this question in some detail.
Exploring and defining TAM sequences
To further explore TnpB diversity tnpB DNA sequences of the 107 IS605 subgroup ISfinder entries (Fig. IS200.4B) were more extensively analyzed [36]) with a view to uncovering differences in activities and identifying highly active members. This analysis did not include the 244 IS1341 members which are flanked by typical IS200-IS605 family secondary structures but carry only a TnpB gene.
Firstly, the IS605 subgroup members were used as a seed to search the non-redundant NCBI nucleotide sequence database. Full length copies were extracted and their flanking sequences were examined to eliminate identical insertion events.
To confirm the ISfinder validation, the right end of each multicopy IS was aligned and the tetranucleotide which forms CR and undergoes special base pairing with the tetranucleotide guide sequence (GR) within RE (Fig. IS200.13) was identified, while the single copy IS were examined and compared to their ISfinder annotations. Additionally, the integrity of tnpB was confirmed. This is important because it has been observed that in IS containing tnpA and tnpB, tnpB is often decayed (see He et al., [102]).
It should be noted that these procedures are always undertaken as a matter of course before any IS200/IS605 family entry is made in ISfinder.
The collection was arranged into 64 bins using a 90% identity threshold and these were named after the IS with the highest copy number in each group (Fig. IS200.43). Many of these groups consisted of only single example although several included a few additional examples.
To examine how the sequence identities between CL and TAM (Fig. IS200.44) correlate over the range of IS605 group members in the ISfinder database distributed over the 64 TnpB bins (Fig. IS200.43), activities were tested separately for each of the 64 using a 2 plasmid, TAM depletion assay (Fig. IS200.44 ii) [36].
One plasmid included ~200 nucleotides of the 3’ IS ends including a 20nt abutting “guide” sequence cloned downstream of a tnpB gene which, when expressed together (Fig. IS200.44 ii), are capable of forming the re(ω)RNA complex. The second plasmid consisted of a library with five randomized base pairs (N5) located 5’ to a target sequence recognized by the guide sequence, an assay similar to that used by Karvelis et al., [72] (Fig. IS200.40). Both plasmids were introduced concomitantly into a host cell. Those that carry an N5 sequence susce, ptible to the corresponding re(ω)RNA complex will be depleted and underrepresented in the plasmid population (reduced level of KmR colonies in the population).
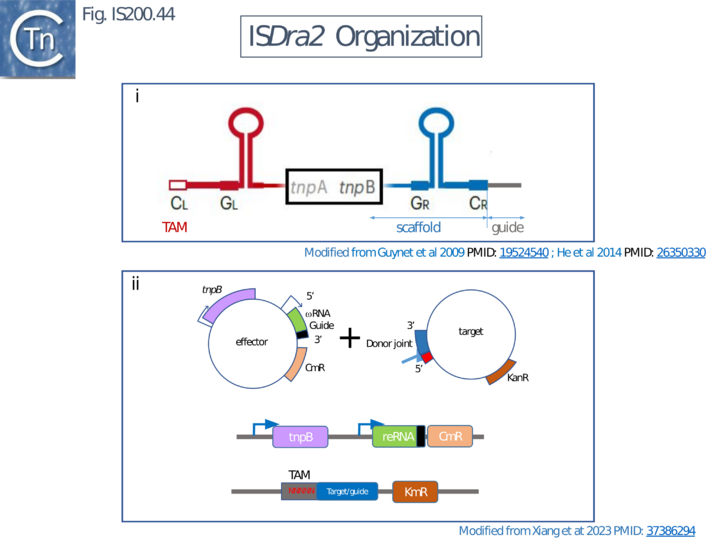
The corresponding TAM sequences (Fig. IS200.43) showed a remarkable identity to the CL sequences with very few variations. For these variants, the authors propose alternative base pairings which would need to be confirmed experimentally.
Further analysis based on a tree generated from TnpB alignments such as those shown in Fig. IS200.35, revealed, perhaps not unexpectedly, that TAM sequences were more similar between closely related IS.
The relative activities of the TAM sequences in each case were then assessed in E. coli using a similar plasmid system to that of Fig. IS200.44, but in which the N5 sequence was substituted for the proposed TAM.
A high proportion (25/64) of these TAM/TnpB derivatives were found to be active.
Sequence requirements of the re(ω)RNA
To explore re(ω)RNA sequence requirements in greater detail, three IS systems, ISTfu1, ISDge10 and ISAba30, in addition to ISDra2, were analyzed in for their guide RNA functions [36].
The relatively small TnpB protein had been demonstrated to function in gene targeting in human cells [72]. Since the interest of Xiang et at [36] was to optimize TnpB as a targeting tool in human cells, the assay was designed for use by transfection into the HEK293T human cell line It used a system in which an out of frame downstream GFP gene was reframed only when the TnpB nuclease could act on its target and the DNA break was repaired by non-homologous joining (Fig. IS200.45 i).
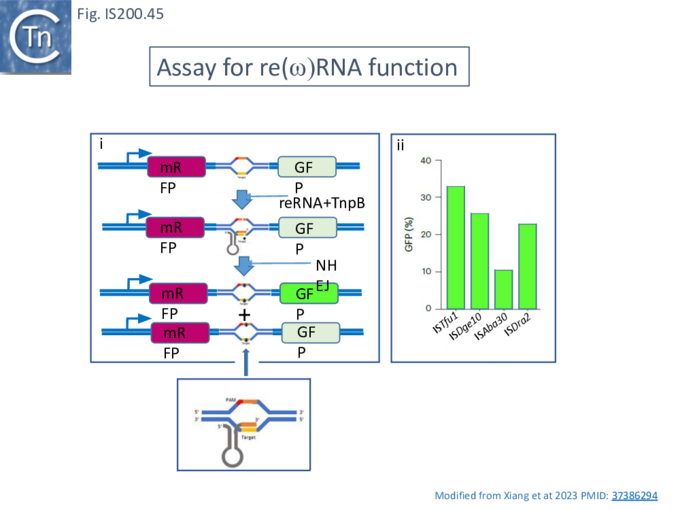
When this reporter plasmid and a TnpB/ re(ω)RNA plasmid were co-transfected, all four TnpB systems were shown to function, yielding 10% to 34% of GFP transfected cells (Fig. IS200.45 ii). They each generated short, deletions of various lengths, some of which lead to placing the GFP gene in phase yielding GFP+ cells in the population. The overall organization of the IS including TAM, scaffold and guide sequence is shown in Fig. IS200.46 i.
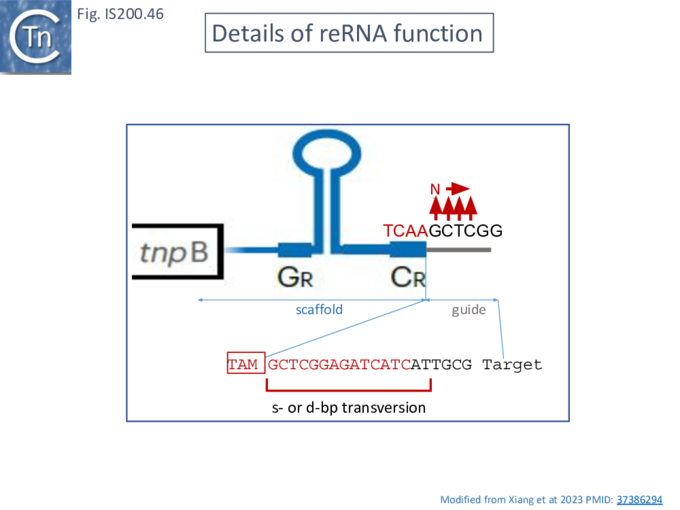
Severely decreased activity in re(ω)RNA guide activity was observed with mutation of either CR or the four proximal nucleotides (Fig. IS200.46) and in the target site with single or double transversion in the TAM proximal region.
It should be noted that where assays were carried out following transfection of human HEK293T cells and it is possible that the results may vary in the appropriate bacterial hosts.
Exploring and defining TAM sequences in a library extracted from NCBI
In a second study to investigate whether the re(ω)RNAs were present across the widely diverse TnpB systems [43] 37272862 constructed a TnpB sequence library, extracted from data from NCBI, which included those associated with Y1 (HUH; IS200-IS605 family), serine (IS607 family) transposases or “non-mobile” orthologues. This generated 5 clades [95]; background in Fig. IS200.47). The clades follow the configuration of the RuvC catalytic motif (Fig. IS200.47) (RuvC-III DRDXN, typical; RuvC-III NADXN, derived) or “catalytic rearrangements (RuvC-II (RII-r3 and 5) or RuvC-III (RIII-r4) domain) [103] (Fig. IS200.47).
The authors chose 59 TnpB orthologs covering the diversity (background to Fig. IS200.47; [95] and varying in length between 353 to 550 aa. The TnpB-re(ω)RNA-encoding loci including a suitable promoter were expressed in an in vitro transcription/translation (IVTT) system and the 5’ ends were determined by RACE from the 3’ re(ω)RNA end lacking the guide sequence.
This identified 30/59 orthologs with a defined 5’ end and lengths of between 79 and 466 nt. TnpBAma generated a 106 nt scaffold, and is thus identical in processing as was found in the experiments of Fig. IS200.41. Some orthologs, such as TnpBDra2 showed multiple 5’ ends, consistent with previous observations suggesting either incomplete or promiscuous RNase activity [72][98].
A screen for DNA nuclease activities of the IVTT-produced re(ω)RNAs revealed that 27/59 were active. They also defined the TAM sequences revealing only limited diversity of these sequences as was also found for the ISfinder collection [36]. The assay was validated by confirming both the AmaTnpBAma (TCAC) and TnpBDra2 (TTGAT) TAM sequences.

re(ω)RNA and tnpB Co-evolution
It was noted that ISDra2 re(ω)RNA includes the 3’ segment of tnpB (residues 335 to 408 and −231G to −10U) which suggests that TnpB and the guide sequence system might have co-evolved [99]. However, although re(ω)RNA expression and processing may require co-expression with the TnpB protein, Nakagawa et al., [98] suggest that co-evolution might be less constrained than previously predicted because, they argue, that functionally essential gene regions and those of re(ω)RNA do not overlap significantly: the structures imply that the TnpB C-terminus (residues 376 to 408 overlapping with −109G to −10U) is not involved in DNA cleavage, and the 5′ re(ω)RNA terminus (−231G to −117T, overlapping with residues 336 to 373) is not required for target DNA cleavage.
The question of co-evolution is complex since it must also take into account the constraints imposed by the mechanism(s) involved in the DNA transposition process: the TAM sequence which abuts the left IS end (LE) also serves as a sequence required for cleavage and insertion at the left end CL and that CL interacts in a complex way with a partially complementary sequence, GL, located at the foot of a stem loop (DNA) structure recognised by the TnpA transposase (see He et al., [102]). Moreover, changing the GL sequence leads to a change in the specificity of insertion – i.e. changes the CL sequence [48]. More importantly, the CR sequence which is an integral part of the IS, plays a central role in both the RNA guide and TnpA-mediated DNA cleavage reactions and interacts both with a sequence at the foot of a secondary structure at the right end (RE), GR. and, in the re(ω)RNA where it forms part of a pseudoknot (Fig. IS200.42) [98] [99].
IscB, like TnpB, is also an RNA-guided Endonuclease
Altae-Tran et al.,[43] also examined a very large number of rather disperse IscB systems for their endonuclease properties, their association with RNA and their capacity as RNA guide proteins. Initial studies concerned a CRISPR associated IscB (marked in the article as Delaware Bay acquatic sample), which when purified from a heterologous Escherichia coli host was associated with an RNA localised directly upstream of iscB which generated a signal in a PAM (TAM) “discovery” assay and was able to generate cleavage products in vitro with the appropriate target.
An alignment of over 500 (non-redundant) iscB genes revealed an upstream region of conserved sequence of about 300 bp which terminated at what the authors state is an IS200/IS605-like end. One specific example examined, present in the host K. racemifer genome in nearly 50 copies, was associated with non-coding RNA species in most cases, which they called ΩRNA, with significant secondary structure potential. An example of K. racemifer IscB was investigated in vitro using a plasmid substrate and shown to: use a target adjacent pentanucleotide TAM, ATAAA; and observed that by changing the complementary RNA extension (guide),cleavage was reprogrammable.
To further characterize IscB, the TAM sequences of 57 examples from a collection of 86 genes from a phylogenetically diverse set of bacteria could be determined; of those 57, 5 were reconstituted with their omega RNA and found to active in target cleavage; and one, AwaIscB from Allochromatium warmingii, was chosen for further study.
Biochemically, IscBAwa could cleave double strand DNA in a magnesium dependent reprogrammable way with a temperature optimum of 35-40°C and with RNA guide lengths of between 15 and 45 nts. A mutation of the RuvC E residue eliminated cleavage of the non-target strand while mutation of an H residues in the HNH motif eliminated cleavage of the target strand (as expected for a Cas9-related enzyme; Fig. IS200.32). Mutation of both residues eliminated cleavage altogether. Also like Cas9, cleavage was: TAM (PAM) proximal (3 nts from TAM for the target and 8 or 12 nts for the non-target strands; that the RNP protected DNA from ExoIII digestion 19 nts upstream of the TAM on the target and 6 downstream on the non-target (Fig. IS200.32); and that truncation of the newly identified N-terminal PLMP domain (named after a cluster of conserved amino acids; Fig. IS200.48 top) eliminated activity.
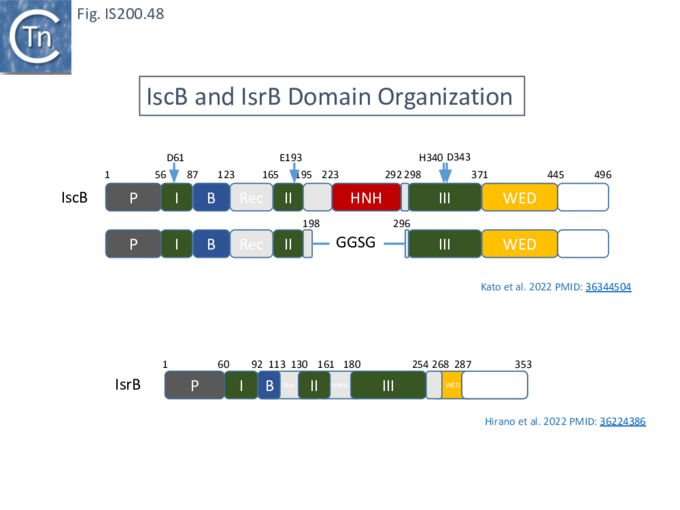
The Structure of IscB–ωRNA ribonucleoprotein complex and the ternary complex containing target DNA.
IscB associates with a 200-400nt ωRNA, significantly longer than the 100nt guide RNA of its probable offspring, Cas9 [43]. IscB are much smaller than Cas9 and lack the α-helical nucleic-acid recognition domain but share the RuvC and HNH endonuclease domains (Fig. IS200.48).
Kato et al., [104] used an IscB protein derived from the human gut metagenome (IscBOgeu) as a model while Hirano et al., [105] used an IrsB (IsrBDt) from Desulfovirgula thermocuniculi. IrsB are related to IscB but lack the HNH nuclease domain (Fig. IS200.48). Note that this is a more detailed description of the domain structure than shown in Fig. IS200.37. A detailed study by Meer et al., [41] found that the IscB and IrsB formed clearly separate groups on a phylogenetic tree.
For the structural cryo-em studies, a catalytically inactivated IscBOgeu E193A (RuvC)/H247A (HNH) derivative was used. In the IscBOgeu structure, the catalytic D61 (RuvC I), E193 (RuvC II), H340, and D343 (RuvC III) and a divalent Mg2+ ion (Fig. IS200.48) are configured similarly to those in Cas9 although the structure lacked the HNH domain.
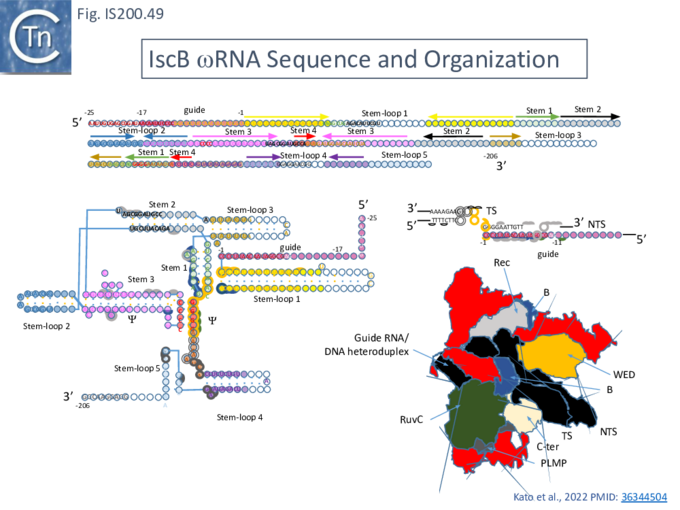
The ωRNA structure is complex (Fig. IS200.49) comprising a 27 nt guide sequence and a 206 nt scaffold with 5 stem loops, 4 stems and a linker. The guide adaptor, stem-loop 1 (yellow), connects the guide segment (dark red) and stem 1 (green; which the authors call the “nexus” stem widely conserved in the tracrRNA of Cas9s; [106]). Stem 1, stem 2 (grey; the central stem), and stem-loop 3 (brown) form a three-way junction. Like TnpB ωRNA, IscBOgeu ωRNA also includes a pseudoknot (??). Stem loop 2 (blue) stacks with the nexus pseudoknot hairpin (pink) which in turn interacts with the pseudoknot stem 4 (red).
The cognate ωRNA and IscBOgeu E193/H247 were expressed in E.coli, the IscB-ωRNA complex purified and the ternary complex assembled by mixing with target DNA. However, to improve resolution, it was found necessary to delete the HNH domain (residues 199 – 295) (Fig. IS200.48), which is flexible in Cas9 [107][108]. The complex, composed of an IscB monomer and a single ωRNA was formed using the deletion derivative IscBω, an ωRNA of 233 nt including a 27 nt guide sequence and a partially double strand DNA target (Fig. IS200.49 right).
In the ternary complex IscB ωRNA guide sequence forms a 14 bp heteroduplex with the target DNA (Fig. IS200.49 middle right) and is recognized by IscB in a sequence-specific fashion using the short Rec region (Fig. IS200.48) shown in grey in Fig. IS200.49 middle right. A simplified cartoon is shown in Fig. IS200.49 bottom right. This is somewhat different from Cas9 which form a 20 bp heteroduplex with a much larger Rec domain. TAM is recognized by the CT domain and mismatches at positions 15 and 16 are tolerated for cleavage. The differences in a full complex with the HNH domain and with the ωHNH IscB derivative is shown in Fig. IS200.50.
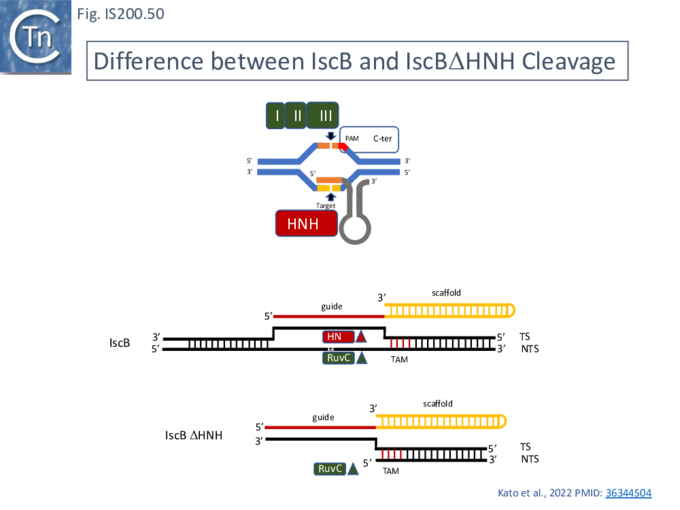
The Structure of IsrB–ωRNA ribonucleoprotein complex and the ternary complex containing target DNA
IsrB is short, about 350 amino acids and lacking an HNH domain (Fig. IS200.51) (therefore equivalent to the ??HNH IscB derivative). It is associated with a long RNA guide of ~300-nt which guides IsrB to nick the non-target strand (NTS) of double-stranded (ds) DNA (see Fig. IS200.51 top) containing a 5′-NTGA-3′ TAM [43].
The Desulfovirgula thermocuniculi IsrB (IsrBDt) ωRNA (284 nt) is longer than that of IscBOgeu, and includes a 20 nt guide segment which forms a heteroduplex with the target DNA [105]. Like IscBOgeu, IsrBDt ωRNA is structurally complex including eight stem loops and four stems (Fig. IS200.51 middle). The structure includes 2 pseudoknots: one defined by two of the stem-loops (2 and 5, red boxes (Fig. IS200.51 middle) and the other the “nexus” pseudoknot (blue boxes).

IsrBDt recognizes the TTGA TAM in the NTS by both hydrogen bonds and van der Waals interactions and cleavage occurred 8–11 nt upstream of TAM, further than the 2–5 nt of Cas9. TAM recognition was more specific at 60 °C for this thermophilic enzyme than at lower temperatures where NTGA was recognized [43].
IsrB diversity of structure and ωRNA architecture
As in numerous publications in this field, Hirano et al., [105], explored IsrB diversity and ωRNA ternary structure. They identified five orthologues and their cognate ωRNAs from: Crocosphaera watsonii (IsrBCw); Dolichospermum sp. (IsrBDs); Calditerricola satsumensis (IsrBCs); Burkholderiales bacterium (IsrBBb); and a viral metagenome assembly (IsrBK2). A standard TAM identification assay (such as that shown in Fig. IS200.40) indicated that IsrBBb recognizes NTGG while IsrBCw, IsrBCs, IsrBDs and IsrBK2 recognize NTG. All were active in an in vitro reconstituted IsrB-ωRNA RNPpromoted nicking of dsDNA substrates.
ωRNAs of the five orthologues and IsrBDs retain the core domain composition: four stems (S1–4) and five stem loops (SL1/2/4/5/7) (Fig. IS200.51 middle). Inspection of the ωRNAs showed some significant architectural differences, however: For example, in a group, including IsrBCs, IsrBK2 and IsrBBb, SL2 and SL4 form pseudoknots, and SL5 and the intermediate region between S2 and SL7 form pseudoknots while in a second group, including IsrBDt, IsrBCw and IsrBDs, SL2 and SL5 form pseudoknots, and SL4 and the intermediate region between S2 and SL7 form pseudoknots.
The IS1341 Conundrum: how do derivatives without their transposase transpose?
IS1341 Group Diversity: Mining the NCBI NR database
Conserved secondary structure motifs
IS1341 group orientation suggests iscB re(Ω)RNA but not tnpB re(Ω)RNA is expressed in transcriptionally active environments.
IS1341 Group Function
Does a Resident TnpA copy Drive IS1341 group Transposition?
TnpBGst and IscBGst proteins are active RNA-guided Nucleases.
TnpB is Required for Replacement of the Deleted IS Copy.
The Copy Choice Model for TnpB Function During Transposition
IStrons
The IS605-based IStron: CdiIStron.
IS607-based IStrons
TnpAS IS607 Excision and Insertion Activity
IStron-encoded TnpB nucleases
Defining the CBoIStron TAM Sequence: a double role in both nuclease and transposase recognition
CBoIStron TnpB/wRNA promotes transposon copy number maintenance
Busy Ends: Functional interactions between IStron splicing, TnpB and ωRNA
Busy Ends
The Eukaryotic Connection: Fanzor eukaryotic TnpB relatives
TnpB Clade
Fanzor1
Fanzor2 and/or Fanzor1 are of bacterial origin
Fanzor2 and/or Fanzor1 may have evolved from an IS607 ancestor
Fanzor1 may have evolved from Fanzor2
Fanzor Activity
Functional Relationship Between Fanzor Evolution and IS607 TnpB
Y1 transposase domestication
TnpAREP and REP/BIME
Acknowledgements
We are grateful to Fred Dyda and Alison Hickman for advice concerning transposition mechanism, to Orsyla Barabas for certain figures and videos of structures, and to Kira Makarova and Virginijus Šikšnys for advice concerning the RNA guide endonucleases. The Siksnys group also kindly supplied the Cas12 structural panel.
Bibliography
- ↑ 1.0 1.1 1.2 1.3 1.4 <pubmed>6313217</pubmed>
- ↑ 2.0 2.1 2.2 2.3 2.4 <pubmed>15179601</pubmed>
- ↑ 3.0 3.1 <pubmed>6315530</pubmed>
- ↑ 4.0 4.1 4.2 <pubmed>3009825</pubmed>
- ↑ 5.0 5.1 <pubmed>9060429</pubmed>
- ↑ <pubmed>2546038</pubmed>
- ↑ <pubmed>8601470</pubmed>
- ↑ 8.0 8.1 <pubmed>7557457</pubmed>
- ↑ 9.0 9.1 9.2 <pubmed>2553665</pubmed>
- ↑ <pubmed>8386127</pubmed>
- ↑ 11.0 11.1 11.2 11.3 11.4 <pubmed>9858724</pubmed>
- ↑ 12.0 12.1 <pubmed>10986230</pubmed>
- ↑ <pubmed>10220167</pubmed>
- ↑ 14.0 14.1 14.2 14.3 14.4 14.5 14.6 <pubmed>11807059</pubmed>
- ↑ 15.0 15.1 <pubmed>26104715</pubmed>
- ↑ <pubmed>26350323</pubmed>
- ↑ 17.0 17.1 <pubmed>23832240</pubmed>
- ↑ <pubmed>9631304</pubmed>
- ↑ 19.0 19.1 19.2 19.3 19.4 19.5 19.6 <pubmed>16209952</pubmed>
- ↑ 20.0 20.1 20.2 20.3 20.4 20.5 20.6 20.7 <pubmed>16163392</pubmed>
- ↑ 21.0 21.1 21.2 21.3 21.4 <pubmed>18280236</pubmed>
- ↑ 22.0 22.1 22.2 22.3 22.4 22.5 22.6 22.7 <pubmed>18243097</pubmed>
- ↑ 23.0 23.1 23.2 23.3 23.4 23.5 23.6 <pubmed>20090938</pubmed>
- ↑ 24.0 24.1 24.2 24.3 24.4 24.5 24.6 <pubmed>20691900</pubmed>
- ↑ 25.0 25.1 25.2 25.3 25.4 25.5 25.6 25.7 25.8 <pubmed>20890269</pubmed>
- ↑ 26.0 26.1 <pubmed>17347521</pubmed>
- ↑ <pubmed>10418150</pubmed>
- ↑ <pubmed>8253675</pubmed>
- ↑ <pubmed>8384142</pubmed>
- ↑ 30.0 30.1 <pubmed>10471738</pubmed>
- ↑ <pubmed>18725932</pubmed>
- ↑ <pubmed>26044710</pubmed>
- ↑ <pubmed>9422611</pubmed>
- ↑ <pubmed>24195768</pubmed>
- ↑ <pubmed>9789049</pubmed>
- ↑ 36.00 36.01 36.02 36.03 36.04 36.05 36.06 36.07 36.08 36.09 36.10 <pubmed>37386294</pubmed>
- ↑ 37.0 37.1 <pubmed>27572647</pubmed>
- ↑ <pubmed>14676423</pubmed>
- ↑ 39.0 39.1 <pubmed>17006450</pubmed>
- ↑ <pubmed>10913072</pubmed>
- ↑ 41.0 41.1 41.2 <pubmed>37758954</pubmed>
- ↑ 42.00 42.01 42.02 42.03 42.04 42.05 42.06 42.07 42.08 42.09 42.10 42.11 42.12 42.13 42.14 42.15 42.16 42.17 <pubmed>PMC4810608</pubmed>
- ↑ 43.00 43.01 43.02 43.03 43.04 43.05 43.06 43.07 43.08 43.09 43.10 43.11 43.12 43.13 43.14 43.15 43.16 43.17 43.18 43.19 43.20 43.21 <pubmed>34591643</pubmed>
- ↑ <pubmed>8374079</pubmed>
- ↑ 45.0 45.1 <pubmed>16340015</pubmed>
- ↑ 46.0 46.1 46.2 46.3 <pubmed>23345619</pubmed>
- ↑ 47.0 47.1 47.2 47.3 47.4 47.5 47.6 47.7 <pubmed>21745812</pubmed>
- ↑ 48.0 48.1 48.2 48.3 <pubmed>19524540</pubmed>
- ↑ <pubmed>29635476</pubmed>
- ↑ 50.0 50.1 <pubmed>16359337</pubmed>
- ↑ <pubmed>19703395</pubmed>
- ↑ <pubmed>9620951</pubmed>
- ↑ <pubmed>3000598</pubmed>
- ↑ <pubmed>2451025</pubmed>
- ↑ <pubmed>2546858</pubmed>
- ↑ <pubmed>21896744</pubmed>
- ↑ <pubmed>1531480</pubmed>
- ↑ <pubmed>1740453</pubmed>
- ↑ <pubmed>12864855</pubmed>
- ↑ <pubmed>27466393</pubmed>
- ↑ <pubmed>649572</pubmed>
- ↑ <pubmed>7309705</pubmed>
- ↑ <pubmed>27298350</pubmed>
- ↑ <pubmed>21479270</pubmed>
- ↑ <pubmed>11753368</pubmed>
- ↑ <pubmed>2838063</pubmed>
- ↑ <pubmed>2553666</pubmed>
- ↑ 68.0 68.1 68.2 <pubmed>23461641</pubmed>
- ↑ <pubmed>24728998</pubmed>
- ↑ <pubmed>PMC5851899</pubmed>
- ↑ <pubmed>31857715</pubmed>
- ↑ 72.00 72.01 72.02 72.03 72.04 72.05 72.06 72.07 72.08 72.09 72.10 72.11 72.12 72.13 72.14 72.15 72.16 72.17 72.18 72.19 72.20 72.21 72.22 72.23 <pubmed>34619744</pubmed>
- ↑ 73.0 73.1 <pubmed>24505130</pubmed>
- ↑ 74.0 74.1 <pubmed>22949671</pubmed>
- ↑ <pubmed>21756346</pubmed>
- ↑ <pubmed>31021231</pubmed>
- ↑ <pubmed>33764415</pubmed>
- ↑ <pubmed>33333018</pubmed>
- ↑ <pubmed>37971304</pubmed>
- ↑ <pubmed>23548000</pubmed>
- ↑ <pubmed>7517937</pubmed>
- ↑ <pubmed>PMC6728339</pubmed>
- ↑ <pubmed>PMC4615843</pubmed>
- ↑ <pubmed>15752202</pubmed>
- ↑ 85.0 85.1 <pubmed>25127548</pubmed>
- ↑ <pubmed>21668986</pubmed>
- ↑ 87.0 87.1 87.2 <pubmed>25806405</pubmed>
- ↑ <pubmed>34209065</pubmed>
- ↑ <pubmed>19536208</pubmed>
- ↑ <pubmed>PMC4247193</pubmed>
- ↑ <pubmed>PMC124278</pubmed>
- ↑ <pubmed>9288893</pubmed>
- ↑ <pubmed>9783582</pubmed>
- ↑ <pubmed>30691644</pubmed>
- ↑ 95.0 95.1 95.2 95.3 95.4 95.5 <pubmed>37272862</pubmed>
- ↑ 96.0 96.1 <pubmed>27096362</pubmed>
- ↑ <pubmed>28431230</pubmed>
- ↑ 98.00 98.01 98.02 98.03 98.04 98.05 98.06 98.07 98.08 98.09 <pubmed>37020030</pubmed>
- ↑ 99.00 99.01 99.02 99.03 99.04 99.05 99.06 99.07 99.08 99.09 <pubmed>37020015</pubmed>
- ↑ <pubmed>23287718</pubmed>
- ↑ <pubmed>26422227</pubmed>
- ↑ 102.0 102.1 102.2 <pubmed>26350330</pubmed>
- ↑ 103.0 103.1 <pubmed>37983496</pubmed>
- ↑ 104.0 104.1 104.2 104.3 104.4 <pubmed>36344504</pubmed>
- ↑ 105.0 105.1 105.2 105.3 <pubmed>36224386</pubmed>
- ↑ <pubmed>25373540</pubmed>
- ↑ <pubmed>29127285</pubmed>
- ↑ <pubmed>26524520</pubmed>


