General Information/IS and Gene Expression: Difference between revisions
m Text replacement - "</pubmed></ref>" to "}}</ref>" |
m Text replacement - "<pubmed>" to "{{#" |
||
| Line 1: | Line 1: | ||
'''<big>A</big>'''nother important aspect of IS impact on their bacterial hosts is their ability to modulate gene expression. In addition to acting as vectors for gene transmission from one replicon to another in the form composite transposons (two IS flanking any gene; [[:Image:1.2.3.png|Fig.2.3]]) and tIS [[:Image:1.13.1.png|(Fig.8.1)]] and their ability to interrupt genes, it has been known for some time<ref>{{#1101028}}</ref><ref name=":0"> | '''<big>A</big>'''nother important aspect of IS impact on their bacterial hosts is their ability to modulate gene expression. In addition to acting as vectors for gene transmission from one replicon to another in the form composite transposons (two IS flanking any gene; [[:Image:1.2.3.png|Fig.2.3]]) and tIS [[:Image:1.13.1.png|(Fig.8.1)]] and their ability to interrupt genes, it has been known for some time<ref>{{#1101028}}</ref><ref name=":0">{{#6271458</pubmed> | ||
</ref> that IS can also activate gene expression. This capacity has recently received much attention due to the increase in resistance to various antibacterials<ref name=":1"> | </ref> that IS can also activate gene expression. This capacity has recently received much attention due to the increase in resistance to various antibacterials<ref name=":1">{{#16952941</pubmed> | ||
</ref><ref name=":2"> | </ref><ref name=":2">{{#12923109</pubmed> | ||
</ref><ref name=":3"> | </ref><ref name=":3">{{#23158541</pubmed> | ||
</ref>, a worrying public health threat<ref>{{#23887414}}</ref><ref>{{#23887415}}</ref>. | </ref>, a worrying public health threat<ref>{{#23887414}}</ref><ref>{{#23887415}}</ref>. | ||
They can accomplish this in two ways: either by providing internal promoters whose transcripts escape into neighboring DNA<ref name=":0" /><ref name=":4"> | They can accomplish this in two ways: either by providing internal promoters whose transcripts escape into neighboring DNA<ref name=":0" /><ref name=":4">{{#6292860</pubmed> | ||
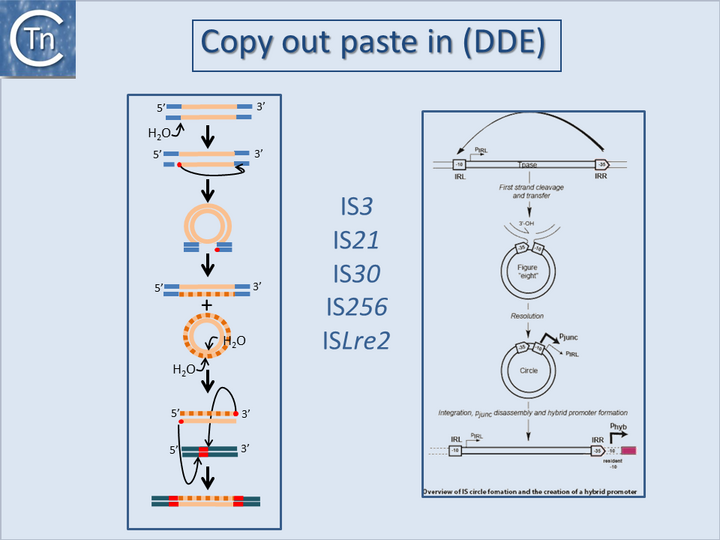
</ref><ref>{{#6260746}}</ref><ref>{{#6311437}}</ref> or by hybrid promoter formation. Many IS carry -35 promoter components oriented towards the flanking DNA [[:Image:1.24.1.png|(Fig.18.1)]]. In a number of cases this plays an important part in their transposition since a significant number of IS transposes using an excised transposon circle [[:Image:1.24.1.png|(Fig.18.1)]] with abutted left and right ends. For these IS, the other end carries a -10 element oriented inwards towards the Tpase gene. Together with the -35, this generates a strong promoter on formation of the circle junction to drive Tpase expression required for catalysis of integration [[:Image:1.24.2.png|(Fig.18.2)]] <ref>{{#26350305}}</ref><ref>{{#9214651}}</ref><ref>{{#10438765}}</ref><ref>{{#11598022}}</ref>. Thus, if integration occurs next to a resident -10 sequence, the IS -35 sequence can contribute to a hybrid promoter to drive expression of neighboring genes [see <ref name=":5"> | </ref><ref>{{#6260746}}</ref><ref>{{#6311437}}</ref> or by hybrid promoter formation. Many IS carry -35 promoter components oriented towards the flanking DNA [[:Image:1.24.1.png|(Fig.18.1)]]. In a number of cases this plays an important part in their transposition since a significant number of IS transposes using an excised transposon circle [[:Image:1.24.1.png|(Fig.18.1)]] with abutted left and right ends. For these IS, the other end carries a -10 element oriented inwards towards the Tpase gene. Together with the -35, this generates a strong promoter on formation of the circle junction to drive Tpase expression required for catalysis of integration [[:Image:1.24.2.png|(Fig.18.2)]] <ref>{{#26350305}}</ref><ref>{{#9214651}}</ref><ref>{{#10438765}}</ref><ref>{{#11598022}}</ref>. Thus, if integration occurs next to a resident -10 sequence, the IS -35 sequence can contribute to a hybrid promoter to drive expression of neighboring genes [see <ref name=":5">{{#3029382}}</ref>]. At present, this phenomenon had been reported to occur with over 30 different IS in more than 17 bacterial species<ref>{{#17223624}}</ref><ref>{{#24499397}}</ref> ([[General Information/IS and Gene Expression#IS and Gene Expression|Table IS and Gene Expression below]]). Indeed, specific vector plasmids have been designed to identify activating insertions (e.g. <ref>{{#8863735}}</ref>). | ||
IS activity can affect efflux mechanisms resulting in increased resistance: [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] or [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] insertion can up-regulate the [[wikipedia:Efflux_(microbiology)|AcrAB-TolC]] pump in ''[[wikipedia:Salmonella_enterica|Salmonella enterica]]''<ref name=":6"> | IS activity can affect efflux mechanisms resulting in increased resistance: [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] or [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] insertion can up-regulate the [[wikipedia:Efflux_(microbiology)|AcrAB-TolC]] pump in ''[[wikipedia:Salmonella_enterica|Salmonella enterica]]''<ref name=":6">{{#15616308</pubmed> | ||
</ref>; [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] or [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS2 IS''2''] insertion upstream of AcrEF<ref name=":7"> | </ref>; [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] or [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS2 IS''2''] insertion upstream of AcrEF<ref name=":7">{{#11302812</pubmed> | ||
</ref><ref>{{#11274125}}</ref> and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS186 IS''186''] insertional inactivation of the AcrAB repressor, AcrR, in ''[[wikipedia:Escherichia_coli|Escherichia coli]] <ref name=":7" />'', all lead to increased resistance to [[wikipedia:Quinolone_antibiotic|fluoroquinolones]]. Insertional inactivation of specific [[wikipedia:Porin_(protein)|porins]] can also play a significant role<ref>{{#15212803}}</ref>.<center>[[Image:1.24.1.png|thumb|center|720x720px|'''Fig.18.1.''' Copy out, paste in (DDE). '''Left column'''. Illustrates the copy out paste in the transposition mechanism. The transposon is represented as a yellow line. Flanking sequences in the donor molecule are blue. Flanking sequences in the target molecule are green. Red circles indicate 3′OH moieties generated by Tpase-catalyzed hydrolysis at the transposon end(s). Red boxes indicate target DNA flanks that are duplicated on insertion. Top to bottom: Tpase catalyzed cleavage at the 3′ transposon ends using H2O as the nucleophile. Liberated 3′OH attack the opposite end to create a bridged molecule which then undergoes replication (dotted line) to generate a circular transponso copy and regenerate the donor molecule. The circle intermediate undergoes cleavage to generate two 3'OH ends which attack the target DNA in a staggered way, resulting in integration. Repair at each end then generates the direct target repeat characteristic of many transposons. | </ref><ref>{{#11274125}}</ref> and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS186 IS''186''] insertional inactivation of the AcrAB repressor, AcrR, in ''[[wikipedia:Escherichia_coli|Escherichia coli]] <ref name=":7" />'', all lead to increased resistance to [[wikipedia:Quinolone_antibiotic|fluoroquinolones]]. Insertional inactivation of specific [[wikipedia:Porin_(protein)|porins]] can also play a significant role<ref>{{#15212803}}</ref>.<center>[[Image:1.24.1.png|thumb|center|720x720px|'''Fig.18.1.''' Copy out, paste in (DDE). '''Left column'''. Illustrates the copy out paste in the transposition mechanism. The transposon is represented as a yellow line. Flanking sequences in the donor molecule are blue. Flanking sequences in the target molecule are green. Red circles indicate 3′OH moieties generated by Tpase-catalyzed hydrolysis at the transposon end(s). Red boxes indicate target DNA flanks that are duplicated on insertion. Top to bottom: Tpase catalyzed cleavage at the 3′ transposon ends using H2O as the nucleophile. Liberated 3′OH attack the opposite end to create a bridged molecule which then undergoes replication (dotted line) to generate a circular transponso copy and regenerate the donor molecule. The circle intermediate undergoes cleavage to generate two 3'OH ends which attack the target DNA in a staggered way, resulting in integration. Repair at each end then generates the direct target repeat characteristic of many transposons. | ||
| Line 30: | Line 30: | ||
! scope="col" |IS family||IS name||Mechanism||Gene(s) affected||Organism||Reference||Clinical/Experimental | ! scope="col" |IS family||IS name||Mechanism||Gene(s) affected||Organism||Reference||Clinical/Experimental | ||
|- | |- | ||
| rowspan="3" |[[IS Families/IS1 family|IS<i>1</i>]]|| rowspan="3" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1A IS<i>1</i>]||Cointegrate||<i>Em<sup>R</sup></i>|| rowspan="2" |<i>[[wikipedia:Escherichia_coli|Escherichia coli]]</i>||<ref name=":8"> | | rowspan="3" |[[IS Families/IS1 family|IS<i>1</i>]]|| rowspan="3" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1A IS<i>1</i>]||Cointegrate||<i>Em<sup>R</sup></i>|| rowspan="2" |<i>[[wikipedia:Escherichia_coli|Escherichia coli]]</i>||<ref name=":8">{{#6283551}}</ref>||C? | ||
|- | |- | ||
|Copy-paste circles||<i>bla<sub>TEM-1</sub></i>||<ref name=":5" />||E | |Copy-paste circles||<i>bla<sub>TEM-1</sub></i>||<ref name=":5" />||E | ||
| Line 72: | Line 72: | ||
|<i>bla<sub>VEB-1</sub></i>/<i>bla<sub>OXA-48</sub></i>||<i>[[wikipedia:Escherichia_coli|Escherichia coli]]</i>||<ref name=":1" />||C | |<i>bla<sub>VEB-1</sub></i>/<i>bla<sub>OXA-48</sub></i>||<i>[[wikipedia:Escherichia_coli|Escherichia coli]]</i>||<ref name=":1" />||C | ||
|- | |- | ||
|[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPa12 IS<i>Pa12</i>]||—||<i>blaPER-1</i>||<i>[[wikipedia:Salmonella_enterica|Salmonella enterica]]</i>, <i>[[wikipedia:Pseudomonas_aeruginosa|Pseudomonas aeruginosa]]</i>, <i>[[wikipedia:Providencia_stuartii|Providencia stuartii]]</i>, <i>[[wikipedia:Acinetobacter_baumannii|Acinetobacter baumannii]]</i>||<ref name=":9"> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPa12 IS<i>Pa12</i>]||—||<i>blaPER-1</i>||<i>[[wikipedia:Salmonella_enterica|Salmonella enterica]]</i>, <i>[[wikipedia:Pseudomonas_aeruginosa|Pseudomonas aeruginosa]]</i>, <i>[[wikipedia:Providencia_stuartii|Providencia stuartii]]</i>, <i>[[wikipedia:Acinetobacter_baumannii|Acinetobacter baumannii]]</i>||<ref name=":9">{{#12936998}}</ref>||C | ||
|- | |- | ||
| rowspan="2" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAba1 IS<i>Aba1</i>]|| rowspan="2" |—||<i>bla<sub>ampC</sub></i>|| rowspan="2" |<i>[[wikipedia:Acinetobacter_baumannii|Acinetobacter baumannii]]</i>||<ref name=":10"> | | rowspan="2" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAba1 IS<i>Aba1</i>]|| rowspan="2" |—||<i>bla<sub>ampC</sub></i>|| rowspan="2" |<i>[[wikipedia:Acinetobacter_baumannii|Acinetobacter baumannii]]</i>||<ref name=":10">{{#12951337}}</ref><ref>{{#14742218}}</ref><ref>{{#16441449}}</ref>||C | ||
|- | |- | ||
|<i>bla<sub>OXA-51</sub></i>/<i>bla<sub>OXA-23</sub></i>||<ref>{{#16630258}}</ref>||C | |<i>bla<sub>OXA-51</sub></i>/<i>bla<sub>OXA-23</sub></i>||<ref>{{#16630258}}</ref>||C | ||
| Line 81: | Line 81: | ||
|[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS<i>5</i>]|| rowspan="6" |—||<i>EmR</i>||<i>[[wikipedia:Escherichia_coli|Escherichia coli]]</i>||<ref name=":8" />||C? | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS<i>5</i>]|| rowspan="6" |—||<i>EmR</i>||<i>[[wikipedia:Escherichia_coli|Escherichia coli]]</i>||<ref name=":8" />||C? | ||
|- | |- | ||
|[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISFtu2 IS''Ftu2'']||general||<i>[[wikipedia:Francisella_tularensis|Francisella tularensis]]</i>||<ref name=":11"> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISFtu2 IS''Ftu2'']||general||<i>[[wikipedia:Francisella_tularensis|Francisella tularensis]]</i>||<ref name=":11">{{#19749055}}</ref>||Natural isolate | ||
|- | |- | ||
|[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISVa1 IS''Va1'']||(iron uptake)||<i>Vibrio anguilarum</i>||<ref>{{#7568465}}</ref>||Natural isolate | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISVa1 IS''Va1'']||(iron uptake)||<i>Vibrio anguilarum</i>||<ref>{{#7568465}}</ref>||Natural isolate | ||
| Line 87: | Line 87: | ||
|[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1168 IS<i>1168</i>]||<i>nimA</i>, <i>nimB</i>||<i>[[wikipedia:Bacteroides|Bacteroides]]</i> sp.||<ref>{{#8067736}}</ref>||C | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1168 IS<i>1168</i>]||<i>nimA</i>, <i>nimB</i>||<i>[[wikipedia:Bacteroides|Bacteroides]]</i> sp.||<ref>{{#8067736}}</ref>||C | ||
|- | |- | ||
|[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1186 IS<i>1186</i>]||<i>cfiA</i>||<i>[[wikipedia:Bacteroides_fragilis|Bacteroides fragilis]]</i>||<ref name=":12"> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1186 IS<i>1186</i>]||<i>cfiA</i>||<i>[[wikipedia:Bacteroides_fragilis|Bacteroides fragilis]]</i>||<ref name=":12">{{#8057831}}</ref><ref>{{#7545155}}</ref>||C | ||
|- | |- | ||
|[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS402 IS<i>402</i>]||<i>bla</i>||<i>[[wikipedia:Burkholderia_cepacia_complex|Pseudomonas cepacia]]</i>||<ref name=":13"> | |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS402 IS<i>402</i>]||<i>bla</i>||<i>[[wikipedia:Burkholderia_cepacia_complex|Pseudomonas cepacia]]</i>||<ref name=":13">{{#3025189}}</ref>||E | ||
|- | |- | ||
| rowspan="6" |[[IS Families/IS6 family|IS<i>6</i>]] | | rowspan="6" |[[IS Families/IS6 family|IS<i>6</i>]] | ||
| Line 140: | Line 140: | ||
| rowspan="2" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1187 IS<i>1187</i>]|| rowspan="3" |—|| rowspan="2" |<i>cfiA</i>|| rowspan="2" |<i>[[wikipedia:Bacteroides_fragilis|Bacteroides fragilis]]</i>||<ref name=":12" />||E+C | | rowspan="2" |[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1187 IS<i>1187</i>]|| rowspan="3" |—|| rowspan="2" |<i>cfiA</i>|| rowspan="2" |<i>[[wikipedia:Bacteroides_fragilis|Bacteroides fragilis]]</i>||<ref name=":12" />||E+C | ||
|- | |- | ||
|<ref name=":3" /><ref name=":14"> | |<ref name=":3" /><ref name=":14">{{#11344163}}</ref><ref name=":15">{{#12604530</pubmed> | ||
</ref>||C | </ref>||C | ||
Revision as of 20:11, 27 May 2025
Another important aspect of IS impact on their bacterial hosts is their ability to modulate gene expression. In addition to acting as vectors for gene transmission from one replicon to another in the form composite transposons (two IS flanking any gene; Fig.2.3) and tIS (Fig.8.1) and their ability to interrupt genes, it has been known for some time[1][2] that IS can also activate gene expression. This capacity has recently received much attention due to the increase in resistance to various antibacterials[3][4][5], a worrying public health threat[6][7].
They can accomplish this in two ways: either by providing internal promoters whose transcripts escape into neighboring DNA[2][8][9][10] or by hybrid promoter formation. Many IS carry -35 promoter components oriented towards the flanking DNA (Fig.18.1). In a number of cases this plays an important part in their transposition since a significant number of IS transposes using an excised transposon circle (Fig.18.1) with abutted left and right ends. For these IS, the other end carries a -10 element oriented inwards towards the Tpase gene. Together with the -35, this generates a strong promoter on formation of the circle junction to drive Tpase expression required for catalysis of integration (Fig.18.2) [11][12][13][14]. Thus, if integration occurs next to a resident -10 sequence, the IS -35 sequence can contribute to a hybrid promoter to drive expression of neighboring genes [see [15]]. At present, this phenomenon had been reported to occur with over 30 different IS in more than 17 bacterial species[16][17] (Table IS and Gene Expression below). Indeed, specific vector plasmids have been designed to identify activating insertions (e.g. [18]).
IS activity can affect efflux mechanisms resulting in increased resistance: IS1 or IS10 insertion can up-regulate the AcrAB-TolC pump in Salmonella enterica[19]; IS1 or IS2 insertion upstream of AcrEF[20][21] and IS186 insertional inactivation of the AcrAB repressor, AcrR, in Escherichia coli [20], all lead to increased resistance to fluoroquinolones. Insertional inactivation of specific porins can also play a significant role[22].
IS and Gene Expression
Bibliography
- ↑ {{#1101028}}
- ↑ 2.0 2.1 2.2 2.3 {{#6271458</pubmed>
- ↑ 3.0 3.1 {{#16952941</pubmed>
- ↑ 4.0 4.1 {{#12923109</pubmed>
- ↑ 5.0 5.1 5.2 5.3 5.4 {{#23158541</pubmed>
- ↑ {{#23887414}}
- ↑ {{#23887415}}
- ↑ 8.0 8.1 {{#6292860</pubmed>
- ↑ {{#6260746}}
- ↑ {{#6311437}}
- ↑ {{#26350305}}
- ↑ {{#9214651}}
- ↑ {{#10438765}}
- ↑ {{#11598022}}
- ↑ 15.0 15.1 {{#3029382}}
- ↑ {{#17223624}}
- ↑ {{#24499397}}
- ↑ {{#8863735}}
- ↑ 19.0 19.1 19.2 {{#15616308</pubmed>
- ↑ 20.0 20.1 20.2 {{#11302812</pubmed>
- ↑ {{#11274125}}
- ↑ {{#15212803}}
- ↑ 23.0 23.1 23.2 {{#6283551}}
- ↑ {{#4610339}}
- ↑ {{#339095}}
- ↑ {{#6187472}}
- ↑ {{#22992527}}
- ↑ {{#12867459}}
- ↑ {{#15130120}}
- ↑ {{#24433026}}
- ↑ {{#6289329}}
- ↑ {{#6311437}}
- ↑ {{#6260374}}
- ↑ 34.0 34.1 {{#12936998}}
- ↑ 35.0 35.1 {{#12951337}}
- ↑ {{#14742218}}
- ↑ {{#16441449}}
- ↑ {{#16630258}}
- ↑ 39.0 39.1 {{#19749055}}
- ↑ {{#7568465}}
- ↑ {{#8067736}}
- ↑ 42.0 42.1 {{#8057831}}
- ↑ {{#7545155}}
- ↑ 44.0 44.1 44.2 {{#3025189}}
- ↑ {{#7840551}}
- ↑ {{#10852863}}
- ↑ {{#18443121}}
- ↑ {{#2160941}}
- ↑ {{#10223953}}
- ↑ {{#6318050}}
- ↑ {{#7517394}}
- ↑ {{#18227185}}
- ↑ {{#3039299}}
- ↑ {{#9756793}}
- ↑ {{#23014718}}
- ↑ {{#3038844}}
- ↑ {{#9371438}}
- ↑ {{#12511511}}
- ↑ {{#9098071}}
- ↑ {{#7830550}}
- ↑ {{#27047473}}
- ↑ 62.0 62.1 62.2 62.3 {{#11344163}}
- ↑ 63.0 63.1 63.2 63.3 {{#12604530</pubmed>
- ↑ {{#8602160}}
- ↑ {{#19744834}}
- ↑ {{#11470367}}
- ↑ {{#12435670}}
- ↑ {{#16569841}}
- ↑ {{#16940134}}
- ↑ {{#9202480}}
- ↑ {{#9765560}}
- ↑ {{#20520730}}