IS Families/IS200 IS605 family
Historical
One of the founding members of this group, IS200, was identified in Salmonella typhimurium [1] as a mutation in hisD (hisD984) which mapped as a point mutation but which did not revert and was polar on the downstream hisC gene (see [2]). S. typhimurium LT2 was found to contain six IS200 copies and the IS was unique to Salmonella [3]. Further studies [4] showed that the IS did not carry repeated sequences, either direct or inverted, at its ends, and that removal of 50 bp at the transposase proximal end (which includes a structure resembling a transcription terminator) removed the strong transcriptional block. IS200 elements from S. typhimurium and S. abortusovis revealed a highly conserved structure of 707–708 bp with a single open-reading-frame potentially encoding a 151 aa peptide and a putative upstream ribosome-binding-site [5].
It has been suggested that a combination of inefficient transcription, protection from impinging transcription by a transcriptional terminator, and repression of translation by a stem-loop mRNA structure. All contribute to tight repression of transposase synthesis [6]. However, although IS200 seems to be relatively inactive in transposition [7], it is involved in chromosome arrangements in S. typhimurium by recombination between copies [8].
A second group of “founding” members of this family was, arguably, IS1341 from the thermophilic bacterium PS3 [9], IS891 from Anabaena sp. M-131 [10] and IS1136 from Saccharopolyspora erythraea [11]. The “transposases” of both elements were observed to be associated in a single IS, IS605, from the gastric pathogen Helicobacter pylori [12]. It was identified in many independent isolates of H. pylori and is now considered to be a central member which defines this large family. IS605 was shown to possess unique, not inverted repeat, ends; did not duplicate target sequences during transposition; and inserted with its left (IS200-homolog) end abutting 5'-TTTAA or 5'-TTTAAC target sequences [13]. Additionally, a second derivative, IS606, with only 25% amino acid identity in the two proteins (orfA and orfB) was also identified in many of the H. pylori isolates including some which were devoid of IS605. The Berg lab also identified another H. pylori IS, IS607 [14] which carried a similar IS1341-like orf (orfB) but with another upstream orf with similarities to that of the mycobacterial IS1535 [15] annotated as a resolvase due the presence of a site-specific serine recombinase motif. Another IS605 derivative, ISHp608, which appeared widely distributed in H. pylori was shown to transpose in E. coli, required only orfA to transpose and inserted downstream from a 5’-TTAC target sequence [16].
General
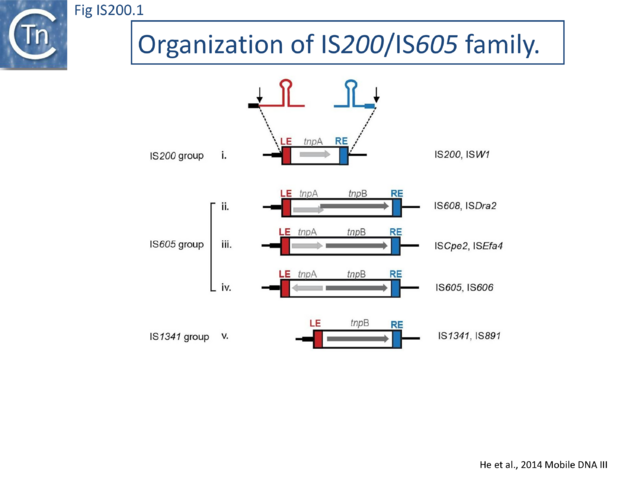
The IS200/IS605 family members transpose using obligatory single strand(ss) DNA intermediates[17] by a mechanism called “peel and paste”. They differ fundamentally in the organization from classical IS. They have sub-terminal palindromic structures rather than terminal IRs (Fig. IS200.1) and insert 3’ to specific AT-rich tetra- or penta-nucleotides without duplicating the target site.
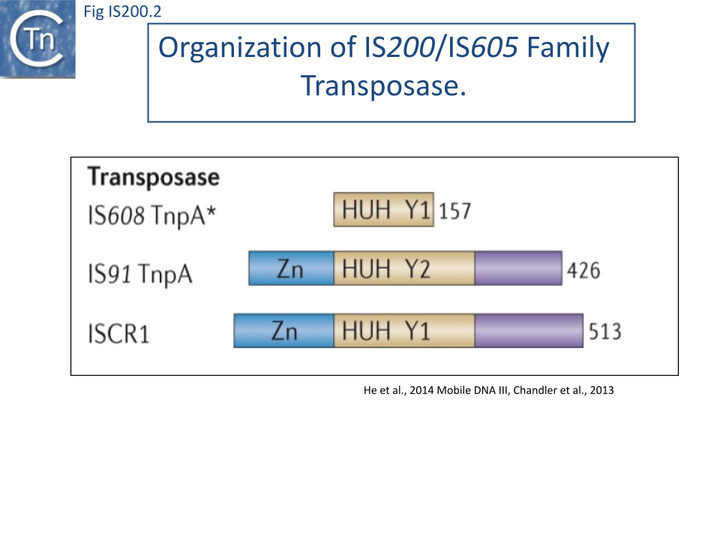
The transposase, TnpA, is a member of the HUH enzyme superfamily (Relaxases, Rep proteins of RCR plasmids/ss phages, bacterial and eukaryotic transposases of IS91/ISCR and Helitrons[18][19])(Fig. IS200.2) which all catalyze cleavage and rejoining of ssDNA substrates.
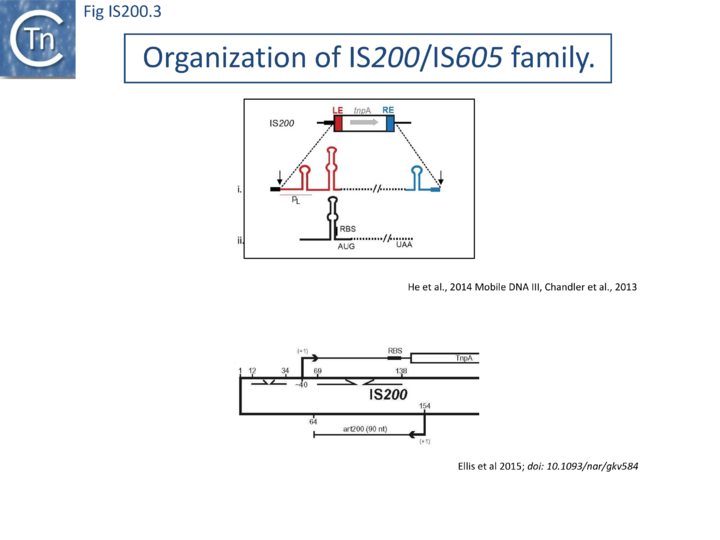
IS200, the founding member (Fig. IS200.3), was identified 30 years ago in Salmonella typhimurium[20] but there has been renewed interest for these elements since the identification of the IS605 group in Helicobacter pylori[21][22][23]. Studies of two elements of this group, IS608 from H. pylori and ISDra2 from the radiation resistant Deinococcus radiodurans, have provided a detailed picture of their mobility [24][25][26][27][28][29][30].
Distribution and Organization
The IS200 group
The IS605 group
The IS1341 group
IS decay
ISC: A group of Elements Related to the IS605 Group
Mechanism of IS200/IS605 single strand DNA transposition
Early models
General transposition pathway
TnpA, Y1 transposases and transposition chemistry
TnpA overall structure
The Single strand Transpososome
Substrate recognition
Cleavage site recognition
Active site assembly and Catalytic activation
Transpososome assembly and stability
Transposition cycle: the trans/cis rotational model
Regulation of single strand transposition
Single strand DNA in vivo
Replication fork
Genome re-assembly after irradiation in D. radiodurans
Real-time transposition (excision) activity
TnpB and its Relatives
IS200/IS605 and the ISC group
ISC have very similar transposases to those of the IS200/IS605 family and are therefore part of the same super family
TnpB, IscB, Cas12 and Cas9
TnpB and IscB are Related to the RNA-guided nucleases Cas12 and Cas9
IscB and Cas9
TnpB and Cas12
Proposed Evolution of TnpB and IscB from an Ancestral RuvC.
Functional analysis of TnpB and IscB
TnpB functions as an RNA-guided Endonuclease
ncRNAs, sotRNAs and reRNAs
TnpB: mechanism of action
An explanation of the “inhibitory effect reported for TnpB?
A system which functions in Eukaryotes
RNA Nomenclature
Generating re(ω)RNA: Processing
The Structure of TnpB-reRNA in association with DNA
TnpB-re(Ω)RNA: Diversity and Activity
Sequence requirements of the re(Ω)RNA
Exploring and defining TAM sequences in a library extracted from NCBI
re(ω)RNA and tnpB Co-evolution
IscB, like TnpB, also functions as an RNA-guided Endonuclease
The Structure of IscB –ωRNA ribonucleoprotein complex and the ternary complex containing target DNA
The Structure of IsrB–ωRNA ribonucleoprotein complex and the ternary complex containing target DNA
IsrB diversity of structure and ωRNA architecture
The IS1341 Conundrum: how do derivatives without their transposase transpose?
IS1341 Group Diversity: Mining the NCBI NR database
Conserved secondary structure motifs
IS1341 group orientation suggests iscB re(Ω)RNA but not tnpB re(Ω)RNA is expressed in transcriptionally active environments.
IS1341 Group Function
Does a Resident TnpA copy Drive IS1341 group Transposition?
TnpBGst and IscBGst proteins are active RNA-guided Nucleases.
TnpB is Required for Replacement of the Deleted IS Copy.
The Copy Choice Model for TnpB Function During Transposition
IStrons
The IS605-based IStron: CdiIStron.
IS607-based IStrons
TnpAS IS607 Excision and Insertion Activity
IStron-encoded TnpB nucleases
Defining the CBoIStron TAM Sequence: a double role in both nuclease and transposase recognition.
CBoIStron TnpB/wRNA promotes transposon maintenance avoiding transposition-associated transposon loss.
Busy Ends: Functional interactions between IStron splicing, TnpB and ωRNA.
Busy Ends
The Eukaryotic Connection: Fanzor eukaryotic TnpB relatives
TnpB Clade
Fanzor1
Fanzor2 and/or Fanzor1 are of bacterial origin
Fanzor2 and/or Fanzor1 may have evolved from an IS607 ancestor
Fanzor1 may have evolved from Fanzor2
Fanzor Activity
The Functional Relationship Between Fanzor Evolution and IS607 TnpB
Y1 transposase domestication
TnpAREP and REP/BIME
Acknowledgements
We are grateful to Fred Dyda and Alison Hickman for advice concerning transposition mechanism, to Orsyla Barabas for certain figures and videos of structures, and to Kira Makarova and Virginijus Šikšnys for advice concerning the RNA guide endonucleases. The Siksnys group also kindly supplied the Cas12 structural panel.
Bibliography
- ↑ <pubmed>6313217</pubmed>
- ↑ <pubmed>15179601</pubmed>
- ↑ <pubmed>6315530</pubmed>
- ↑ <pubmed>3009825</pubmed>
- ↑ <pubmed>9060429</pubmed>
- ↑ <pubmed>15179601</pubmed>
- ↑ <pubmed>2546038</pubmed>
- ↑ <pubmed>8601470</pubmed>
- ↑ <pubmed>7557457</pubmed>
- ↑ <pubmed>2553665</pubmed>
- ↑ <pubmed>8386127</pubmed>
- ↑ <pubmed>9858724</pubmed>
- ↑ <pubmed>9858724</pubmed>
- ↑ <pubmed>10986230</pubmed>
- ↑ <pubmed>10220167</pubmed>
- ↑ <pubmed>11807059</pubmed>
- ↑ <pubmed>26104715</pubmed>
- ↑ <pubmed>26350323</pubmed>
- ↑ <pubmed>23832240</pubmed>
- ↑ <pubmed>6313217</pubmed>
- ↑ <pubmed>9858724</pubmed>
- ↑ <pubmed>9631304</pubmed>
- ↑ <pubmed>11807059</pubmed>
- ↑ <pubmed>16209952</pubmed>
- ↑ <pubmed>16163392</pubmed>
- ↑ <pubmed>18280236</pubmed>
- ↑ <pubmed>18243097</pubmed>
- ↑ <pubmed>20090938</pubmed>
- ↑ <pubmed>20691900</pubmed>
- ↑ <pubmed>20890269</pubmed>