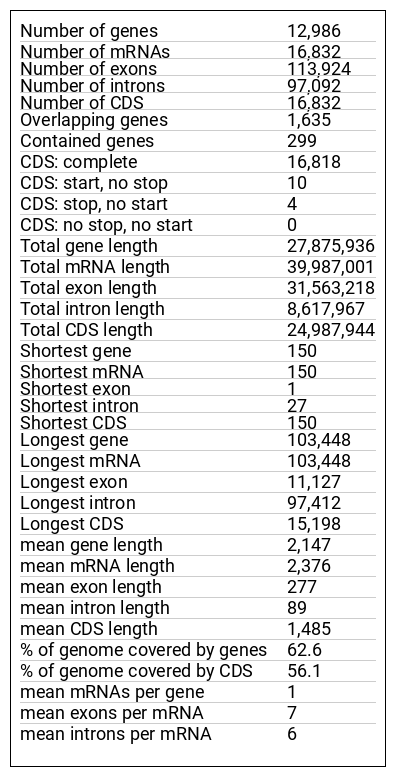
Genome Browser Content and Instructions
jBrowser2 is a platform for visualizing genomic data. The A. subrufescens genome browser contains the following tracks: a) the gene models accompanied by annotation integrated in InterProScan visualization of each coding region; b) Transposable Elements and Repeats Annotation; c) PASA assemblies; d) tRNA and rRNA annotation
;
and e) Aligned public available RNAseq data from different tissues (PRJNA305463
): fruiting-body, mycelium, and primordium.
Use the "open track selector" to navigate into the different and avaliable tracks.